5 Diagnostic Plots
- read in the csv datasets:
- Residuals
library(PKPDmisc)
library(knitr)
library(tidyverse)
#> Loading tidyverse: ggplot2
#> Loading tidyverse: tibble
#> Loading tidyverse: tidyr
#> Loading tidyverse: readr
#> Loading tidyverse: purrr
#> Loading tidyverse: dplyr
#> Conflicts with tidy packages ----------------------------------------------
#> filter(): dplyr, stats
#> lag(): dplyr, statsresid <- read_csv("../data/Residuals.csv")
#> Parsed with column specification:
#> cols(
#> Scenario = col_character(),
#> ID = col_integer(),
#> IVAR = col_double(),
#> TAD = col_double(),
#> PRED = col_double(),
#> IPRED = col_double(),
#> DV = col_double(),
#> IRES = col_double(),
#> Weight = col_double(),
#> IWRES = col_double(),
#> WRES = col_double(),
#> CWRES = col_double(),
#> CdfDV = col_integer(),
#> TADSeq = col_integer(),
#> ObsName = col_character(),
#> ResetSeq = col_integer()
#> )- Create a Res vs Time function with loess fits for the central tendency and the spread (hint abs() is your friend for the spread).
- Conditionally allow the loess curve of central tendency to appear, with a default of TRUE.
- Users should be able to specify the residual column name.
gg_res_tad <- function(df, .tad, .res, .show_loess = TRUE) {
.tad <- rlang::enexpr(.tad)
.res <- rlang::enexpr(.res)
ple <- rlang::quo(
df %>%
ggplot(aes(x = !!.tad, y = !!.res)) + geom_point() +
stat_smooth(data = df %>%
mutate(!!.res := abs(!!.res)),
se = F, color = "blue") +
stat_smooth(data = df %>%
mutate(!!.res := -abs(!!.res)),
se = F, color = "blue") +
theme_bw()
)
output <- rlang::eval_tidy(ple)
if (.show_loess) {
return(
output +
stat_smooth(method = "loess", se=F, color = "red")
)
}
return(output)
}5.0.1 CWRES vs time after dose (TAD)
gg_res_tad(resid, TAD, CWRES)
#> `geom_smooth()` using method = 'loess'
#> `geom_smooth()` using method = 'loess'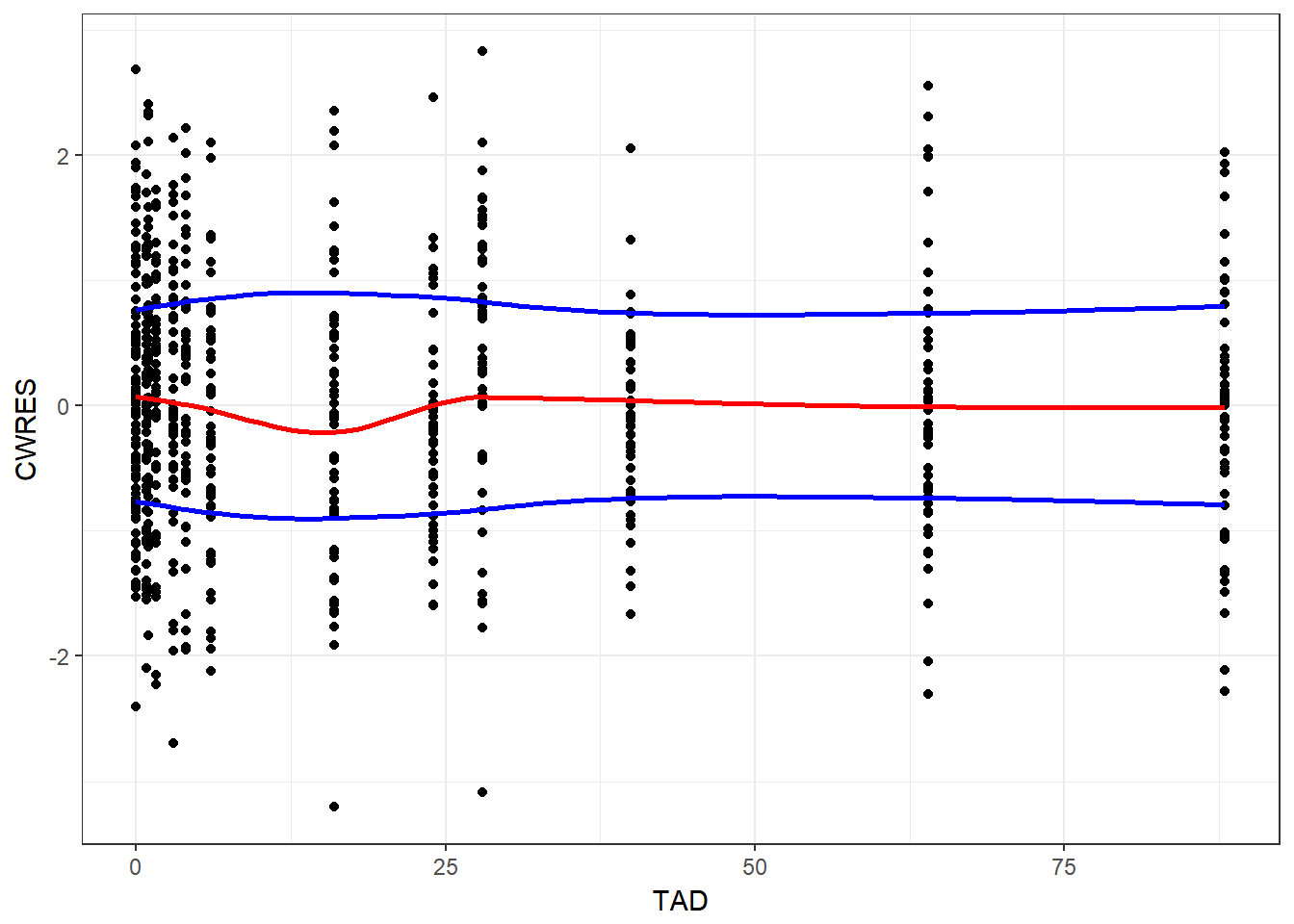
- update your function to flag any point over some threshold as red, with a default of absolute difference of > 2.5
gg_res_tad <- function(df, .tad, .res, .threshold = 2.5, .show_loess = TRUE) {
.tad <- rlang::enexpr(.tad)
.res <- rlang::enexpr(.res)
ple <- rlang::quo(
df %>%
mutate(HIGHRES__ = ifelse(abs(!!.res) > .threshold, 1, 0)) %>%
ggplot(aes(x = !!.tad, y = !!.res)) +
geom_point(aes(color = factor(HIGHRES__))) +
scale_color_manual(values = c("black", "red"),
name = "Outlier",
labels = c("not outlier", "outlier")) +
stat_smooth(data = df %>%
mutate(!!.res := abs(!!.res)),
se = F, color = "blue") +
stat_smooth(data = df %>%
mutate(!!.res := -abs(!!.res)),
se = F, color = "blue") +
theme_bw()
)
output <- rlang::eval_tidy(ple)
if (.show_loess) {
return(
output +
stat_smooth(method = "loess", se=F, color = "red")
)
}
return(output)
}gg_res_tad(resid, TAD, CWRES)
#> `geom_smooth()` using method = 'loess'
#> `geom_smooth()` using method = 'loess'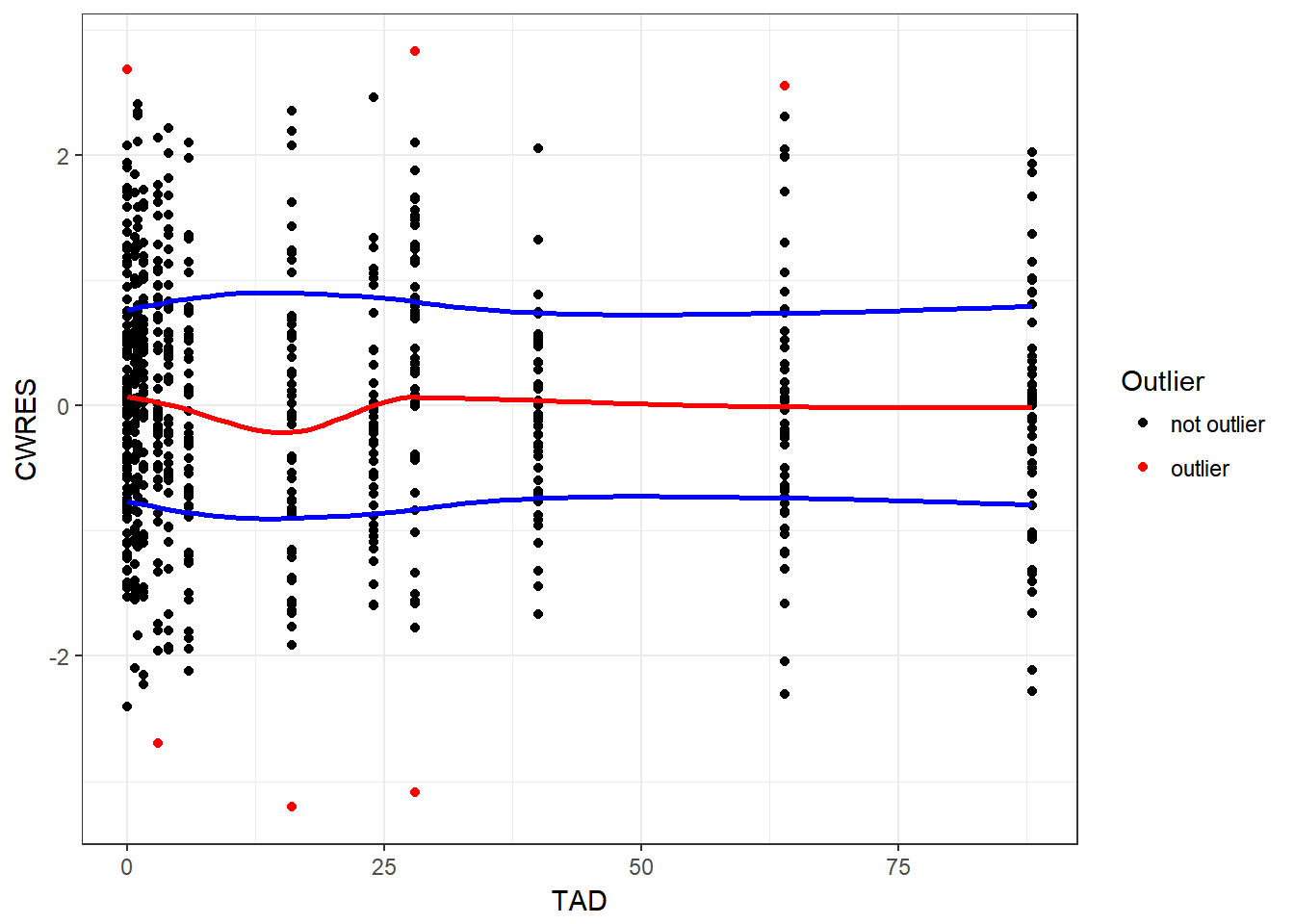
devtools::session_info()
#> Session info -------------------------------------------------------------
#> setting value
#> version R version 3.4.0 (2017-04-21)
#> system x86_64, mingw32
#> ui RTerm
#> language (EN)
#> collate English_United States.1252
#> tz Europe/Prague
#> date 2017-06-05
#> Packages -----------------------------------------------------------------
#> package * version date source
#> assertthat 0.2.0 2017-04-11 CRAN (R 3.4.0)
#> backports 1.1.0 2017-05-22 CRAN (R 3.4.0)
#> base * 3.4.0 2017-04-21 local
#> bindr 0.1 2016-11-13 CRAN (R 3.4.0)
#> bindrcpp * 0.1 2016-12-11 CRAN (R 3.4.0)
#> bookdown 0.4 2017-05-20 CRAN (R 3.4.0)
#> broom 0.4.2 2017-02-13 CRAN (R 3.4.0)
#> cellranger 1.1.0 2016-07-27 CRAN (R 3.4.0)
#> codetools 0.2-15 2016-10-05 CRAN (R 3.4.0)
#> colorspace 1.3-2 2016-12-14 CRAN (R 3.4.0)
#> compiler 3.4.0 2017-04-21 local
#> datasets * 3.4.0 2017-04-21 local
#> devtools 1.13.1 2017-05-13 CRAN (R 3.4.0)
#> digest 0.6.12 2017-01-27 CRAN (R 3.4.0)
#> dplyr * 0.6.0 2017-06-02 Github (tidyverse/dplyr@b064c4b)
#> evaluate 0.10 2016-10-11 CRAN (R 3.4.0)
#> forcats 0.2.0 2017-01-23 CRAN (R 3.4.0)
#> foreign 0.8-67 2016-09-13 CRAN (R 3.4.0)
#> ggplot2 * 2.2.1 2016-12-30 CRAN (R 3.4.0)
#> glue 1.0.0 2017-04-17 CRAN (R 3.4.0)
#> graphics * 3.4.0 2017-04-21 local
#> grDevices * 3.4.0 2017-04-21 local
#> grid 3.4.0 2017-04-21 local
#> gtable 0.2.0 2016-02-26 CRAN (R 3.4.0)
#> haven 1.0.0 2016-09-23 CRAN (R 3.4.0)
#> hms 0.3 2016-11-22 CRAN (R 3.4.0)
#> htmltools 0.3.6 2017-04-28 CRAN (R 3.4.0)
#> httr 1.2.1 2016-07-03 CRAN (R 3.4.0)
#> jsonlite 1.5 2017-06-01 CRAN (R 3.4.0)
#> knitr * 1.16 2017-05-18 CRAN (R 3.4.0)
#> labeling 0.3 2014-08-23 CRAN (R 3.4.0)
#> lattice 0.20-35 2017-03-25 CRAN (R 3.4.0)
#> lazyeval 0.2.0 2016-06-12 CRAN (R 3.4.0)
#> lubridate 1.6.0 2016-09-13 CRAN (R 3.4.0)
#> magrittr 1.5 2014-11-22 CRAN (R 3.4.0)
#> memoise 1.1.0 2017-04-21 CRAN (R 3.4.0)
#> methods 3.4.0 2017-04-21 local
#> mnormt 1.5-5 2016-10-15 CRAN (R 3.4.0)
#> modelr 0.1.0 2016-08-31 CRAN (R 3.4.0)
#> munsell 0.4.3 2016-02-13 CRAN (R 3.4.0)
#> nlme 3.1-131 2017-02-06 CRAN (R 3.4.0)
#> parallel 3.4.0 2017-04-21 local
#> PKPDmisc * 1.0.0 2017-06-02 Github (dpastoor/PKPDmisc@23e1f49)
#> plyr 1.8.4 2016-06-08 CRAN (R 3.4.0)
#> psych 1.7.5 2017-05-03 CRAN (R 3.4.0)
#> purrr * 0.2.2.2 2017-05-11 CRAN (R 3.4.0)
#> R6 2.2.1 2017-05-10 CRAN (R 3.4.0)
#> Rcpp 0.12.11 2017-05-22 CRAN (R 3.4.0)
#> readr * 1.1.1 2017-05-16 CRAN (R 3.4.0)
#> readxl 1.0.0 2017-04-18 CRAN (R 3.4.0)
#> reshape2 1.4.2 2016-10-22 CRAN (R 3.4.0)
#> rlang 0.1.1 2017-05-18 CRAN (R 3.4.0)
#> rmarkdown 1.5.9000 2017-06-03 Github (rstudio/rmarkdown@ea515ef)
#> rprojroot 1.2 2017-01-16 CRAN (R 3.4.0)
#> rvest 0.3.2 2016-06-17 CRAN (R 3.4.0)
#> scales 0.4.1 2016-11-09 CRAN (R 3.4.0)
#> stats * 3.4.0 2017-04-21 local
#> stringi 1.1.5 2017-04-07 CRAN (R 3.4.0)
#> stringr 1.2.0 2017-02-18 CRAN (R 3.4.0)
#> tibble * 1.3.3 2017-05-28 CRAN (R 3.4.0)
#> tidyr * 0.6.3 2017-05-15 CRAN (R 3.4.0)
#> tidyverse * 1.1.1 2017-01-27 CRAN (R 3.4.0)
#> tools 3.4.0 2017-04-21 local
#> utils * 3.4.0 2017-04-21 local
#> withr 1.0.2 2016-06-20 CRAN (R 3.4.0)
#> xml2 1.1.1 2017-01-24 CRAN (R 3.4.0)
#> yaml 2.1.14 2016-11-12 CRAN (R 3.4.0)