1 Introduction to ggplot2
library(PKPDmisc)
library(PKPDdatasets)
library(tidyverse)
#> Loading tidyverse: ggplot2
#> Loading tidyverse: tibble
#> Loading tidyverse: tidyr
#> Loading tidyverse: readr
#> Loading tidyverse: purrr
#> Loading tidyverse: dplyr
#> Conflicts with tidy packages ----------------------------------------------
#> filter(): dplyr, stats
#> lag(): dplyr, stats1.1 XY Plots
- For concentration-time plots filter only OCC 1 from dapa_IV_oral (in PKPDdatasets) for subsequent plotting
data1 <- dapa_IV_oral %>% filter(OCC == 1)- Basic Concentration-Time plot (point and lines)
data1 %>% ggplot(aes(x = TIME, y = COBS, group = ID)) +
geom_point() + geom_line()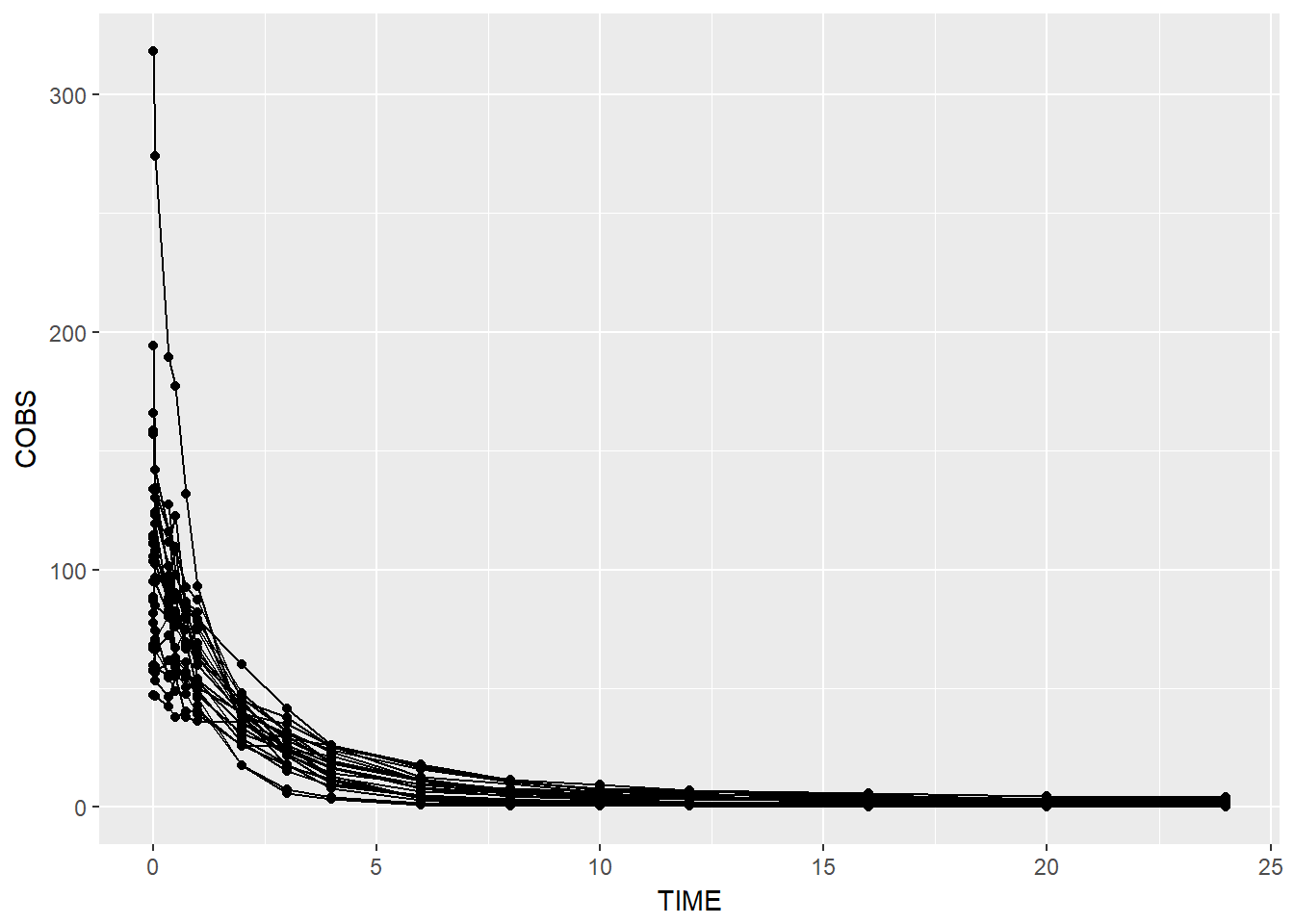
- make points/lines bigger
data1 %>% ggplot(aes(x = TIME, y = COBS, group = ID)) +
geom_point(size = 2.5) + geom_line(size = 1.1)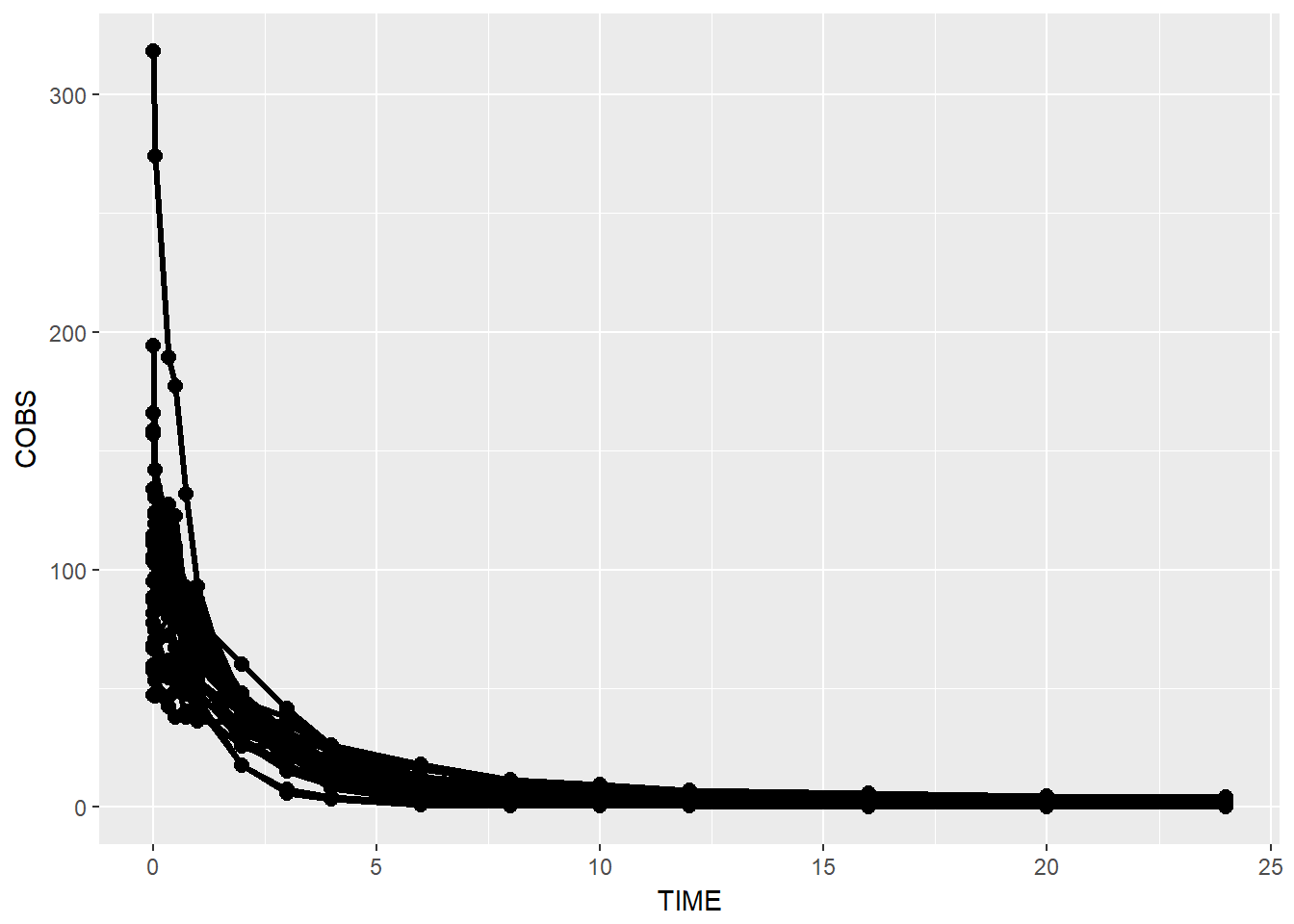
- add log transformation to y axis
data1 %>% ggplot(aes(x = TIME, y = COBS, group = ID)) +
geom_point(size = 2.5) + geom_line(size = 1.1) +
scale_y_log10()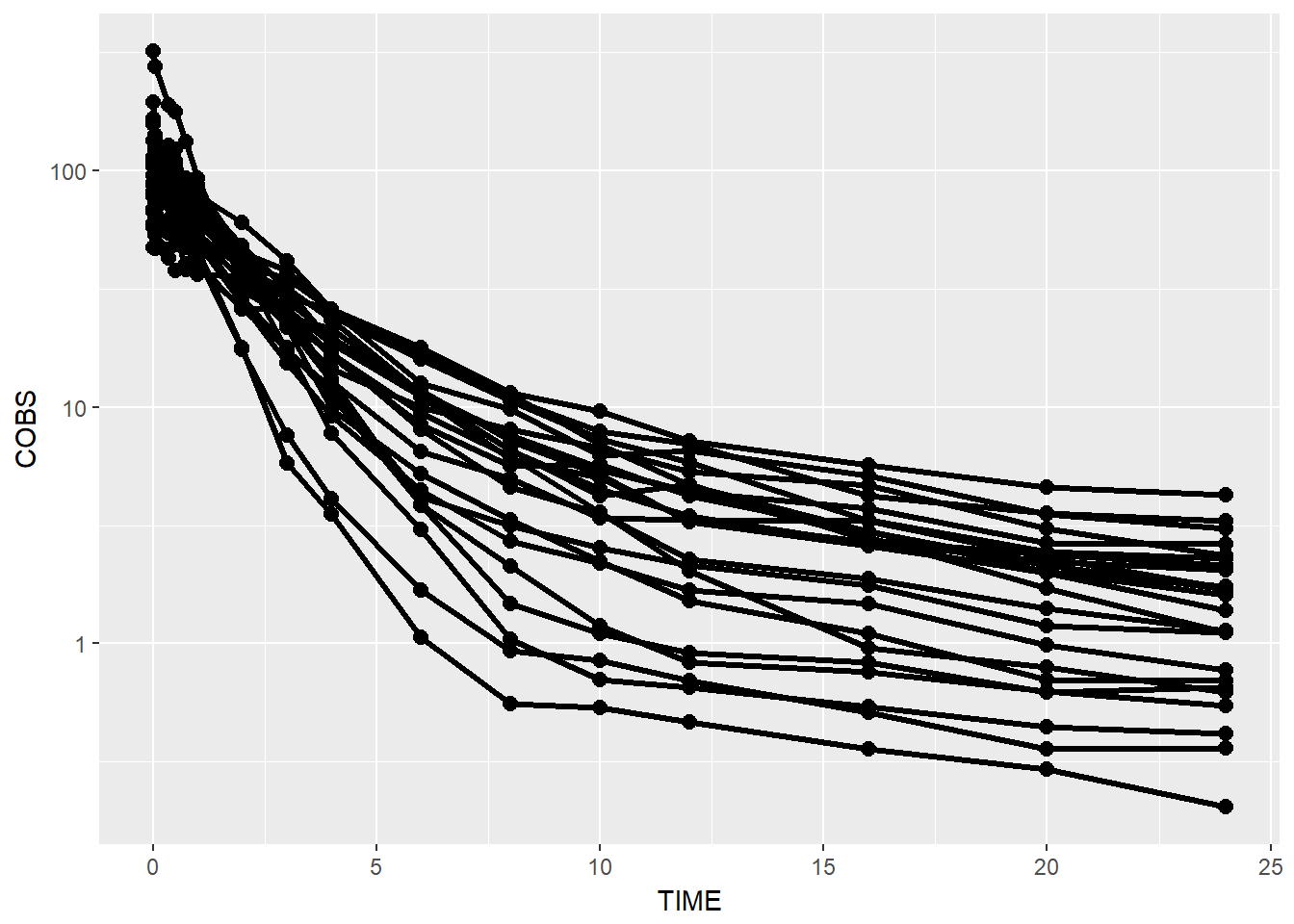
- Add color by gender
data1 %>%
ggplot(aes(x = TIME, y = COBS, group = ID, color = GENDER)) +
geom_point(size = 2.5) + geom_line(size = 1.1) +
scale_y_log10()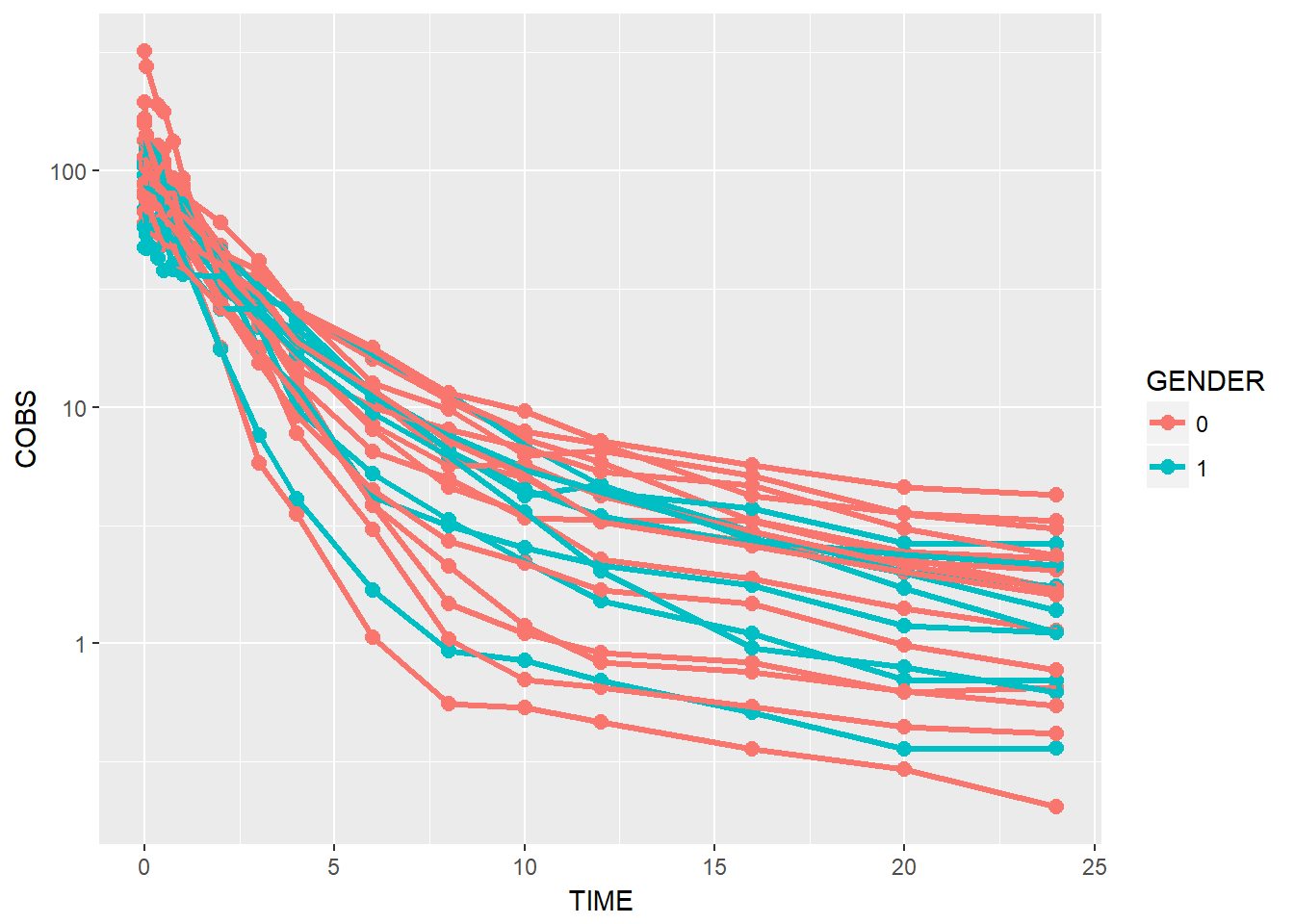
BONUS: rename labels legend
data1 %>%
ggplot(aes(x = TIME, y = COBS, group = ID, color = GENDER)) +
geom_point(size = 2.5) + geom_line(size = 1.1) +
scale_y_log10() + scale_color_discrete(labels = c("Male", "Female"))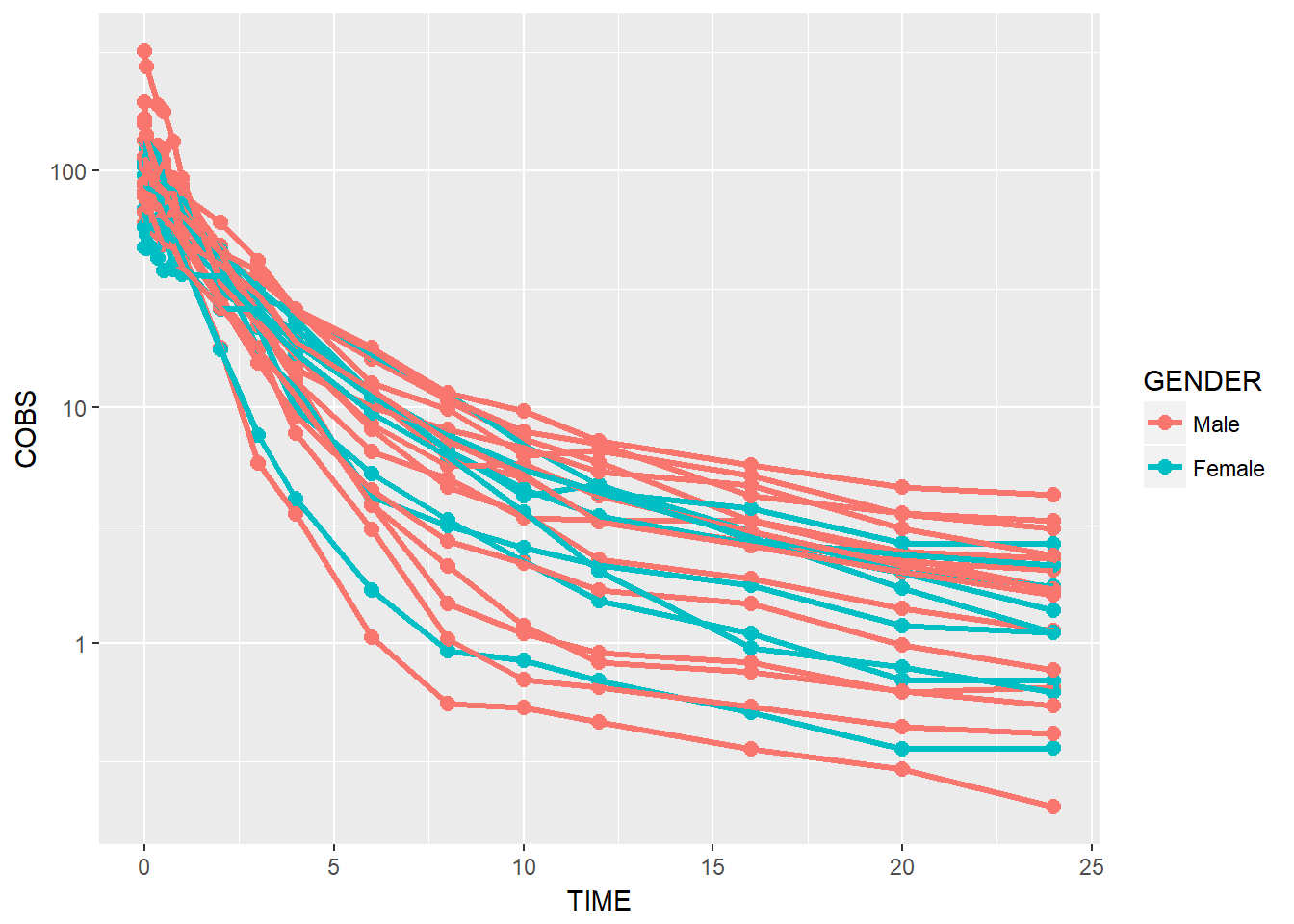
BONUS: move legend to top right
data1 %>%
ggplot(aes(x = TIME, y = COBS, group = ID, color = GENDER)) +
geom_point(size = 2.5) + geom_line(size = 1.1) +
scale_y_log10() + scale_color_discrete(labels = c("Male", "Female")) +
theme(legend.position = c(1, 1), legend.justification = c(1, 1))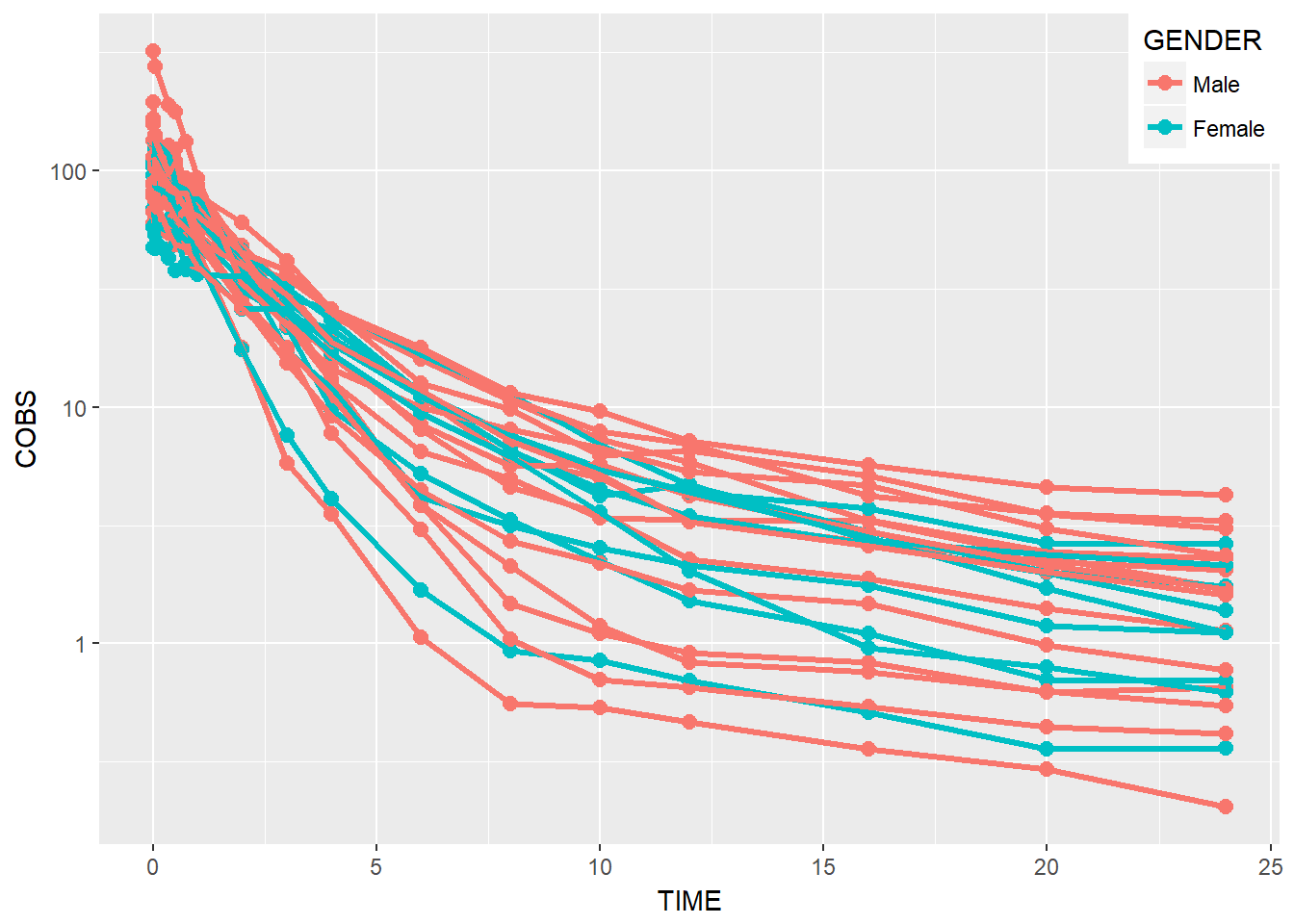
- facet by race
Note, with facetting by race, the color becomes unnessary extra visual stimulus so we remove the color.
data1 %>%
ggplot(aes(x = TIME, y = COBS, group = ID)) +
geom_point(size = 2.5) + geom_line(size = 1.1) +
scale_y_log10() +
theme(legend.position = c(1, 1), legend.justification = c(1, 1)) +
facet_wrap(~GENDER)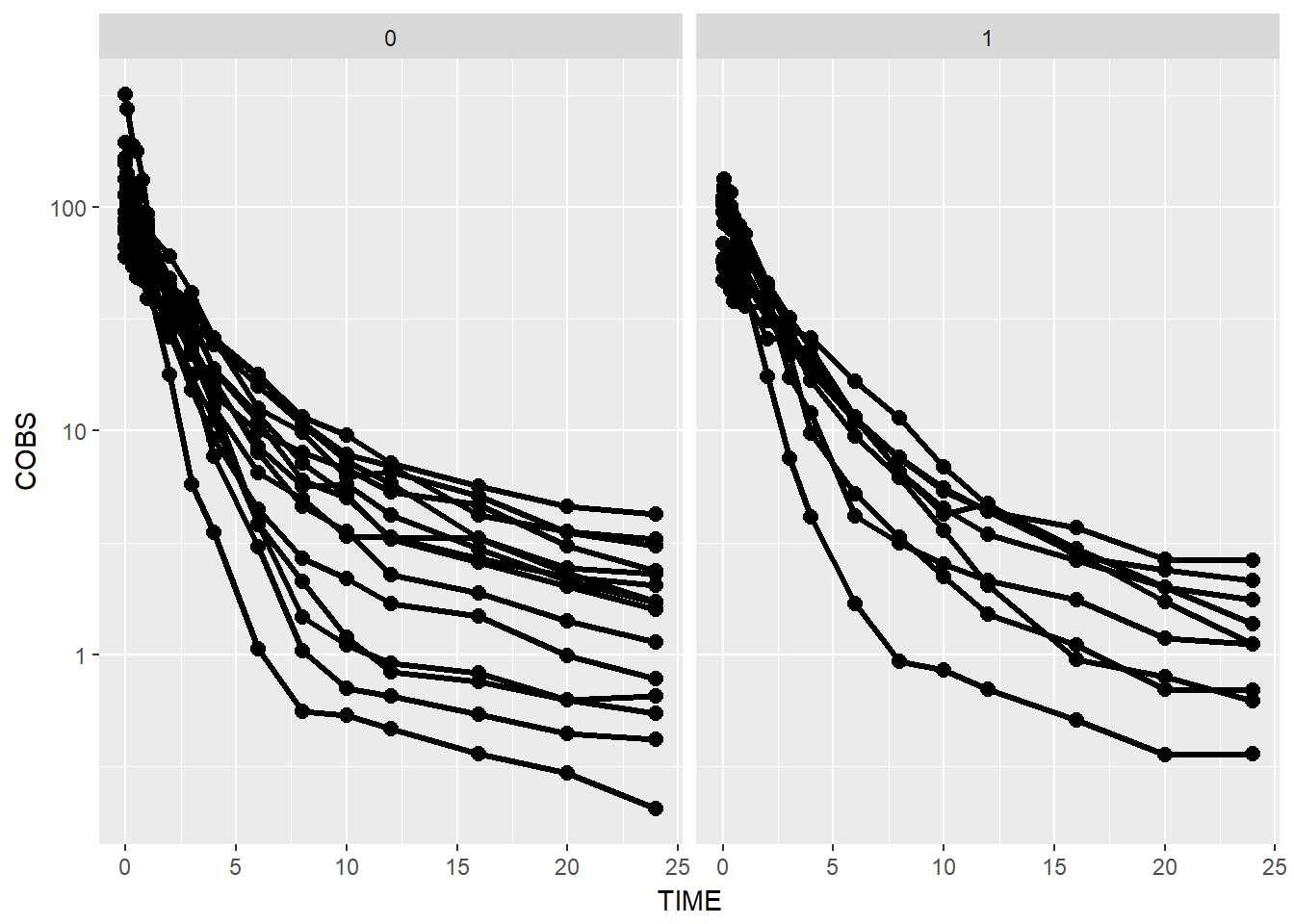
BONUS: rename facet strips by name
There are a couple ways of doing this:
- on the fly mutate the the values to labels
# this is the only time you will see assigning to a column directly
# in this workshop, we encourage to always use mutate to manage columns
# in a dataframe, however as it has not been formally introduced we're
# using base R here
data1$GENDER <- factor(data1$GENDER,
levels = c(0, 1),
labels = c("Male", "Female"))
data1 %>%
ggplot(aes(x = TIME, y = COBS, group = ID)) +
geom_point(size = 2.5) + geom_line(size = 1.1) +
scale_y_log10() +
theme(legend.position = c(1, 1), legend.justification = c(1, 1)) +
facet_wrap(~GENDER)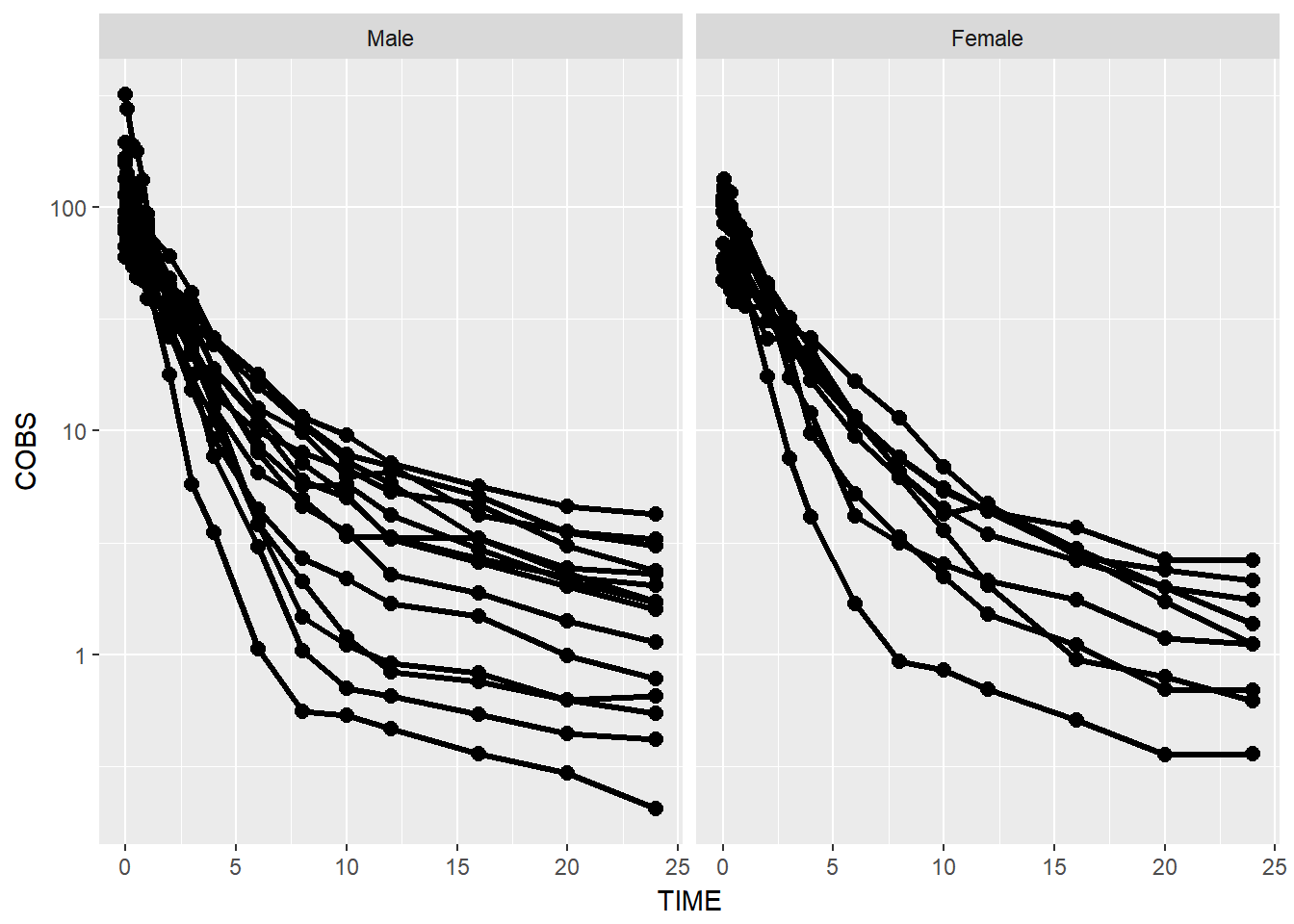
Use facet_grid with the labeller argument
data1 %>%
ggplot(aes(x = TIME, y = COBS, group = ID)) +
geom_point(size = 2.5) + geom_line(size = 1.1) +
scale_y_log10() +
theme(legend.position = c(1, 1), legend.justification = c(1, 1)) +
facet_grid(.~GENDER, labeller = label_both)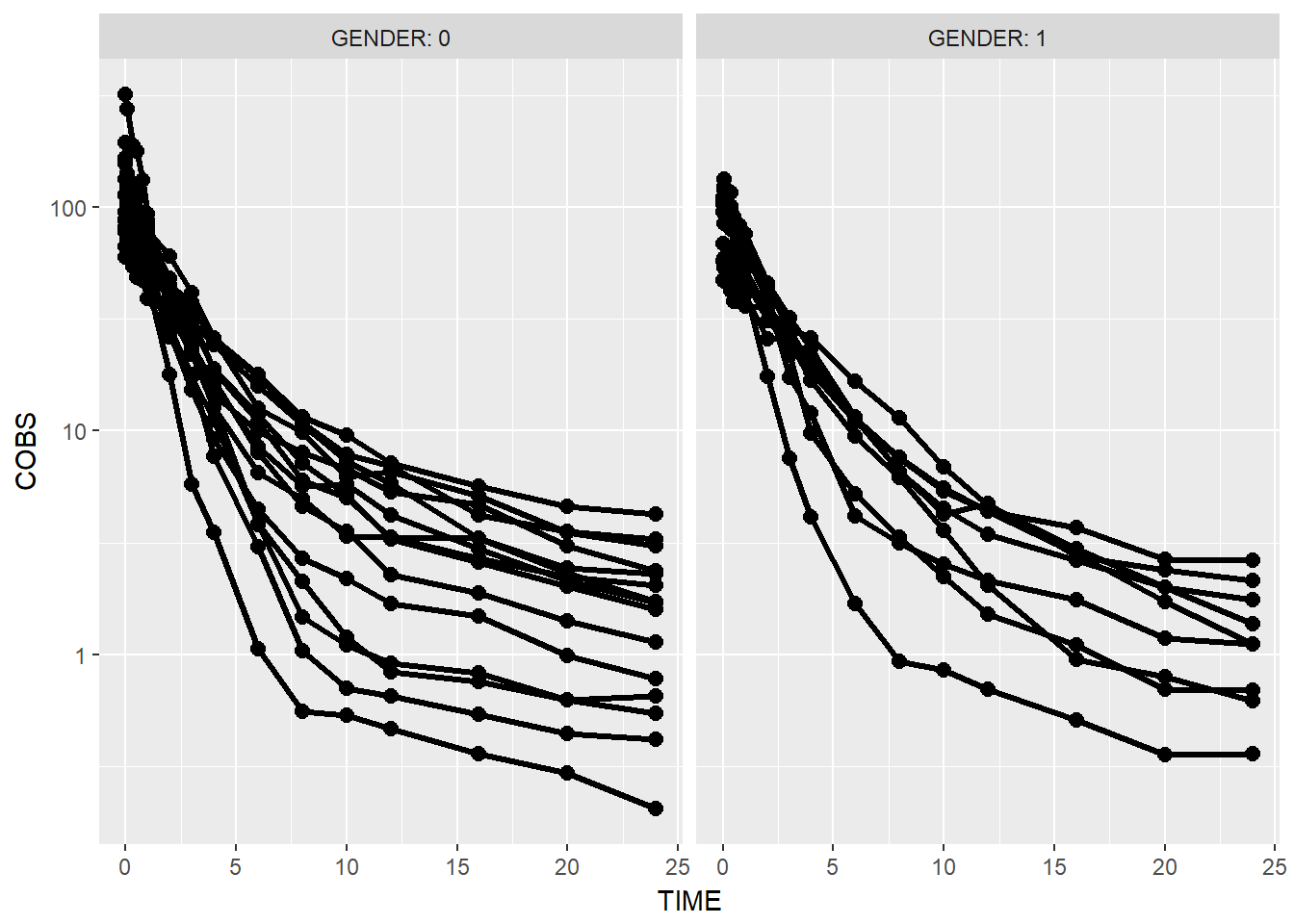
- color by weight
data1 %>%
ggplot(aes(x = TIME, y = COBS, group = ID, color = WEIGHT)) +
geom_point(size = 2.5) + geom_line(size = 1.1) +
scale_y_log10() +
theme(legend.position = c(1, 1), legend.justification = c(1, 1))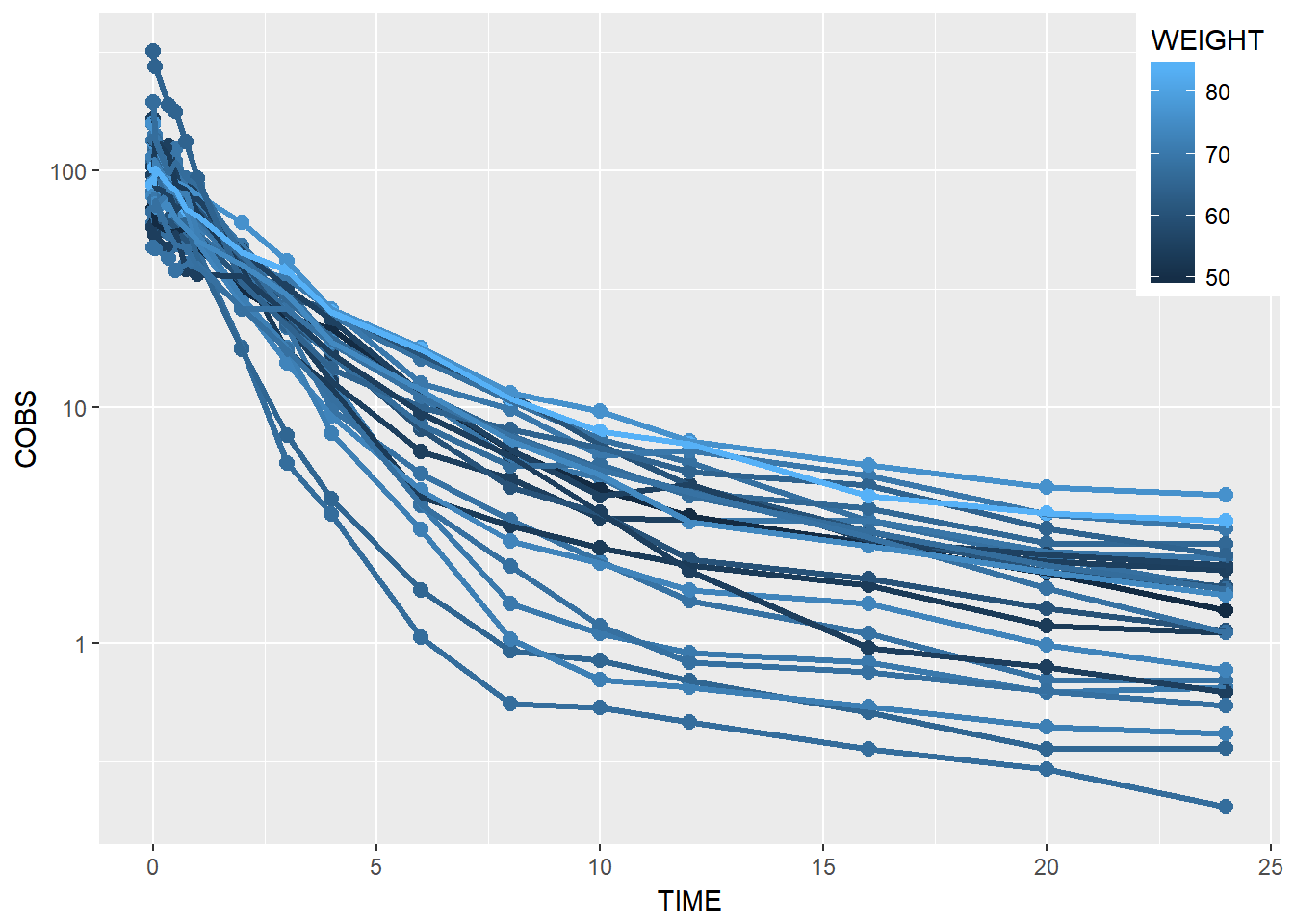
BONUS: rename axes
data1 %>%
ggplot(aes(x = TIME, y = COBS, group = ID, color = WEIGHT)) +
geom_point(size = 2.5) + geom_line(size = 1.1) +
scale_y_log10() +
theme(legend.position = c(1, 1),
legend.justification = c(1, 1)) +
labs(x = "Time, hours", y = "Concentration, ug/mL")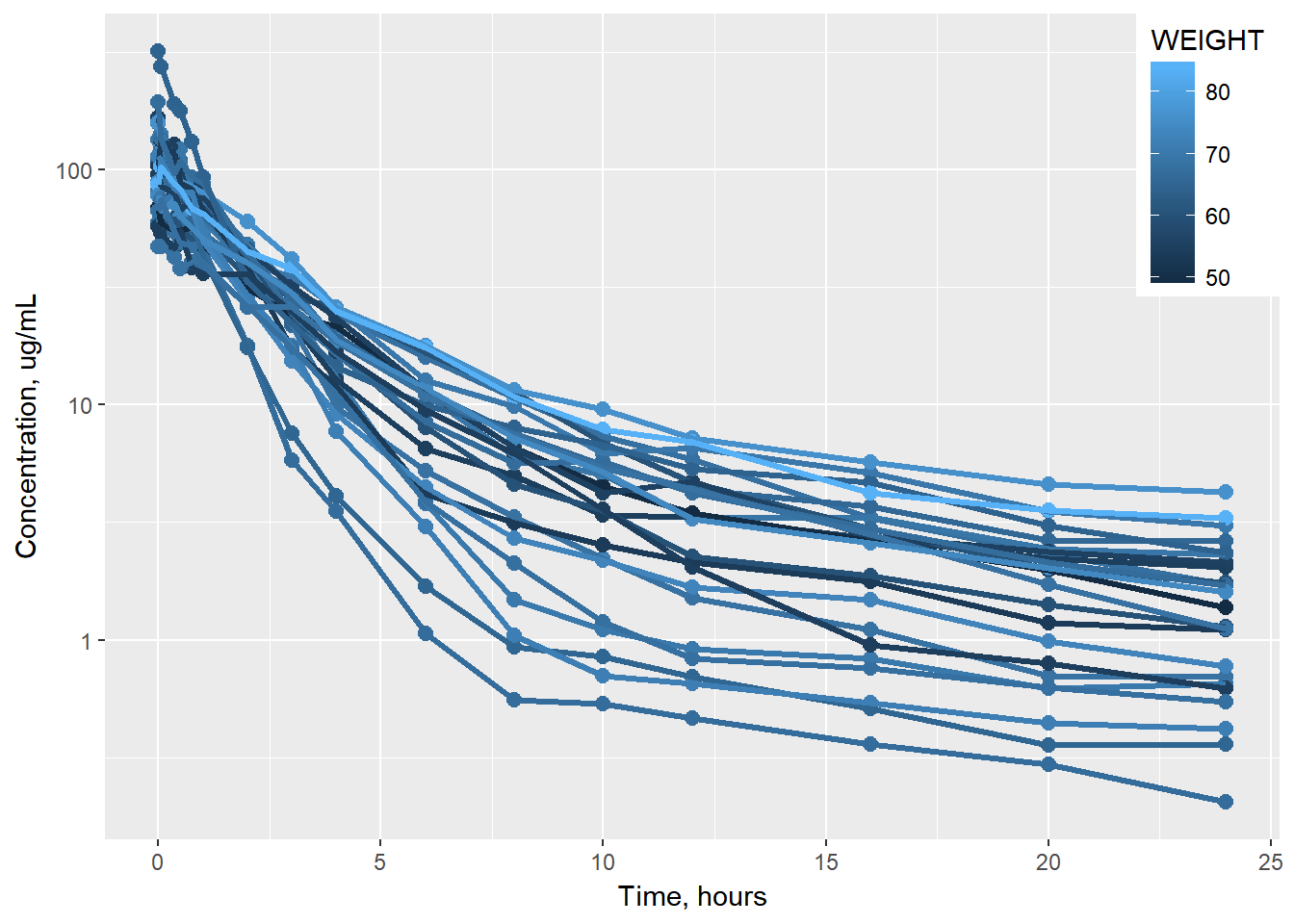
1.2 Boxplots and Histograms
- Histogram(s) of demographics
# distinct is a dplyr verb - easiest way to subset data by the
# first row in some combination.
sid_data <- data1 %>%
distinct(ID, .keep_all = TRUE) # single row per id data
sid_data %>%
ggplot(aes(x = WEIGHT)) +
geom_histogram(binwidth= 4, color="black", fill="white")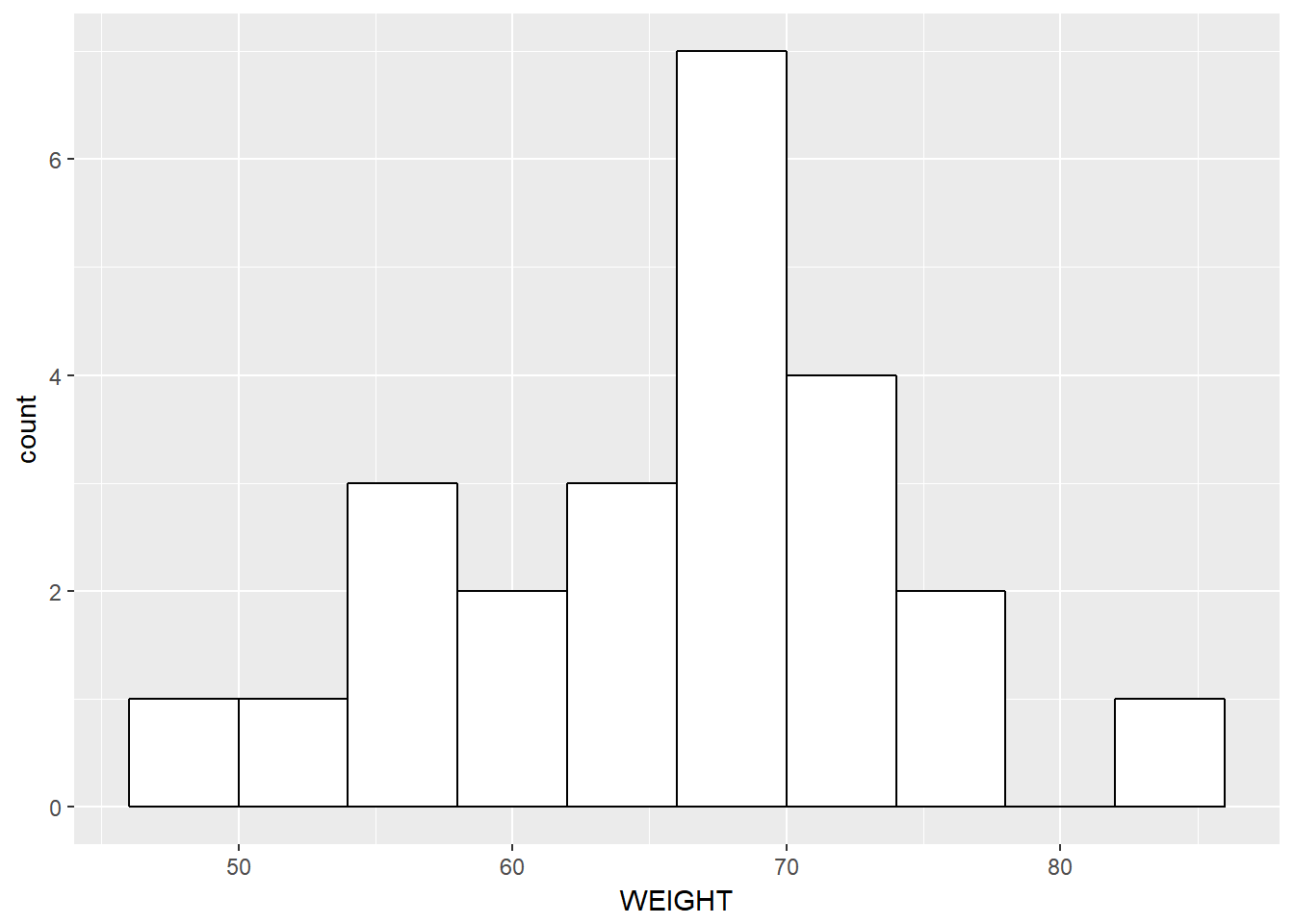
- add vertical line for median value
ggplot(data = sid_data,
aes(x = WEIGHT)) +
geom_histogram(binwidth= 4, color="black", fill="white") +
geom_vline(aes(xintercept = median(WEIGHT)), size= 2, color = "red")
devtools::session_info()
#> Session info -------------------------------------------------------------
#> setting value
#> version R version 3.4.0 (2017-04-21)
#> system x86_64, mingw32
#> ui RTerm
#> language (EN)
#> collate English_United States.1252
#> tz Europe/Prague
#> date 2017-06-05
#> Packages -----------------------------------------------------------------
#> package * version date source
#> assertthat 0.2.0 2017-04-11 CRAN (R 3.4.0)
#> backports 1.1.0 2017-05-22 CRAN (R 3.4.0)
#> base * 3.4.0 2017-04-21 local
#> bindr 0.1 2016-11-13 CRAN (R 3.4.0)
#> bindrcpp * 0.1 2016-12-11 CRAN (R 3.4.0)
#> bookdown 0.4 2017-05-20 CRAN (R 3.4.0)
#> broom 0.4.2 2017-02-13 CRAN (R 3.4.0)
#> cellranger 1.1.0 2016-07-27 CRAN (R 3.4.0)
#> codetools 0.2-15 2016-10-05 CRAN (R 3.4.0)
#> colorspace 1.3-2 2016-12-14 CRAN (R 3.4.0)
#> compiler 3.4.0 2017-04-21 local
#> datasets * 3.4.0 2017-04-21 local
#> devtools 1.13.1 2017-05-13 CRAN (R 3.4.0)
#> digest 0.6.12 2017-01-27 CRAN (R 3.4.0)
#> dplyr * 0.6.0 2017-06-02 Github (tidyverse/dplyr@b064c4b)
#> evaluate 0.10 2016-10-11 CRAN (R 3.4.0)
#> forcats 0.2.0 2017-01-23 CRAN (R 3.4.0)
#> foreign 0.8-67 2016-09-13 CRAN (R 3.4.0)
#> ggplot2 * 2.2.1 2016-12-30 CRAN (R 3.4.0)
#> glue 1.0.0 2017-04-17 CRAN (R 3.4.0)
#> graphics * 3.4.0 2017-04-21 local
#> grDevices * 3.4.0 2017-04-21 local
#> grid 3.4.0 2017-04-21 local
#> gtable 0.2.0 2016-02-26 CRAN (R 3.4.0)
#> haven 1.0.0 2016-09-23 CRAN (R 3.4.0)
#> hms 0.3 2016-11-22 CRAN (R 3.4.0)
#> htmltools 0.3.6 2017-04-28 CRAN (R 3.4.0)
#> httr 1.2.1 2016-07-03 CRAN (R 3.4.0)
#> jsonlite 1.5 2017-06-01 CRAN (R 3.4.0)
#> knitr 1.16 2017-05-18 CRAN (R 3.4.0)
#> labeling 0.3 2014-08-23 CRAN (R 3.4.0)
#> lattice 0.20-35 2017-03-25 CRAN (R 3.4.0)
#> lazyeval 0.2.0 2016-06-12 CRAN (R 3.4.0)
#> lubridate 1.6.0 2016-09-13 CRAN (R 3.4.0)
#> magrittr 1.5 2014-11-22 CRAN (R 3.4.0)
#> memoise 1.1.0 2017-04-21 CRAN (R 3.4.0)
#> methods 3.4.0 2017-04-21 local
#> mnormt 1.5-5 2016-10-15 CRAN (R 3.4.0)
#> modelr 0.1.0 2016-08-31 CRAN (R 3.4.0)
#> munsell 0.4.3 2016-02-13 CRAN (R 3.4.0)
#> nlme 3.1-131 2017-02-06 CRAN (R 3.4.0)
#> parallel 3.4.0 2017-04-21 local
#> PKPDdatasets * 0.1.0 2017-06-02 Github (dpastoor/PKPDdatasets@9eaa831)
#> PKPDmisc * 1.0.0 2017-06-02 Github (dpastoor/PKPDmisc@23e1f49)
#> plyr 1.8.4 2016-06-08 CRAN (R 3.4.0)
#> psych 1.7.5 2017-05-03 CRAN (R 3.4.0)
#> purrr * 0.2.2.2 2017-05-11 CRAN (R 3.4.0)
#> R6 2.2.1 2017-05-10 CRAN (R 3.4.0)
#> Rcpp 0.12.11 2017-05-22 CRAN (R 3.4.0)
#> readr * 1.1.1 2017-05-16 CRAN (R 3.4.0)
#> readxl 1.0.0 2017-04-18 CRAN (R 3.4.0)
#> reshape2 1.4.2 2016-10-22 CRAN (R 3.4.0)
#> rlang 0.1.1 2017-05-18 CRAN (R 3.4.0)
#> rmarkdown 1.5.9000 2017-06-03 Github (rstudio/rmarkdown@ea515ef)
#> rprojroot 1.2 2017-01-16 CRAN (R 3.4.0)
#> rvest 0.3.2 2016-06-17 CRAN (R 3.4.0)
#> scales 0.4.1 2016-11-09 CRAN (R 3.4.0)
#> stats * 3.4.0 2017-04-21 local
#> stringi 1.1.5 2017-04-07 CRAN (R 3.4.0)
#> stringr 1.2.0 2017-02-18 CRAN (R 3.4.0)
#> tibble * 1.3.3 2017-05-28 CRAN (R 3.4.0)
#> tidyr * 0.6.3 2017-05-15 CRAN (R 3.4.0)
#> tidyverse * 1.1.1 2017-01-27 CRAN (R 3.4.0)
#> tools 3.4.0 2017-04-21 local
#> utils * 3.4.0 2017-04-21 local
#> withr 1.0.2 2016-06-20 CRAN (R 3.4.0)
#> xml2 1.1.1 2017-01-24 CRAN (R 3.4.0)
#> yaml 2.1.14 2016-11-12 CRAN (R 3.4.0)