4 Bayesian Problems (and solutions)
Some of the problem areas in bayesian analysis techniques are …..
- autocorrelation
- sensitivity to priors
- …
4.1 Chains
Technique - a ‘master’ execution file can be used to generate new paramater values that can be used for multi-chain runs.
- NSAMPLE - sets number of preturbed parameter estimates, in this case 4
- ISAMPLE - if subsequently estimating, will use the parameters generated from this sample
- by setting to 0 will not tweak these parameters
$EST METHOD=CHAIN FILE=run003chains.chn NSAMPLE=4 ISAMPLE=0 DF=10The larger the DF (degrees of freedom for randomly creating the OMEGAS) the smaller the difference between newly generated values. Eg, with DF=10000 almost no difference in values, whereas DF=5 will have 2x+ differences in values
4.1.0.1 example output comparing output from all chains
** TODO: add more instructions on what all is going on **
set.seed(1234567)
modt <- read_file("../modeling/run006c.modt")
map(1:4, ~ write_file(
infuse(modt,
chain_number = .,
seed = round(runif(1, 1000, 100000), 0)),
file.path("..", "modeling", paste0("run006c", ., ".mod"))))
#> [[1]]
#> $PROB one cmpt simple mu modeled bayes
#> $SUB ADVAN1 TRANS2
#> $INPUT ID TIME DV CMT EVID AMT RATE
#> $DATA mdata/simple_nocovar_50id_6tp.csv IGNORE=@
#> $ABBR REPLACE THETA(CL, V) = THETA(1 to 2)
#> $ABBR REPLACE ETA(CL, V) = ETA(1 to 2)
#>
#> $THETAI
#> THETA(1:NTHETA)=LOG(THETAI(1:NTHETA))
#> THETAP(1:NTHETAP)=LOG(THETAPI(1:NTHETAP))
#> $THETAR
#> THETAR(1:NTHETA)=EXP(THETA(1:NTHETA))
#> THETAPR(1:NTHETAP)=EXP(THETAP(1:NTHETAP))
#>
#> $PRIOR NWPRI
#>
#> $PK
#> "USE NMBAYES_INT, ONLY: ITER_REPORT,BAYES_EXTRA_REQUEST,BAYES_EXTRA
#> ; Request extra information for Bayesian analysis.
#> ; An extra call will then be made for accepted samples
#> "BAYES_EXTRA_REQUEST=1
#>
#> MU_1 = THETA(CL)
#> MU_2 = THETA(V)
#> CL = EXP(MU_1 + ETA(CL))
#> V = EXP(MU_2 + ETA(V))
#> S1 = V
#>
#> "IF(BAYES_EXTRA==1 .AND. ITER_REPORT>=0 .AND. TIME==0.0) THEN
#> "WRITE(50,98) ITER_REPORT,ID,CL,V
#> "98 FORMAT(I12,1X,F14.0,4(1X,1PG12.5))
#> "ENDIF
#>
#> $ERROR
#> "USE NMBAYES_INT, ONLY: ITER_REPORT,BAYES_EXTRA_REQUEST,BAYES_EXTRA
#> "BAYES_EXTRA_REQUEST=1
#> IPRED=F
#> Y = IPRED*(1 + ERR(1))
#> "IF(BAYES_EXTRA==1 .AND. ITER_REPORT>=0 ) THEN
#> "WRITE(51,97) ITER_REPORT,ID,TIME,F
#> "97 FORMAT(I12,1X,F14.0,2(1X,1PG12.5))
#> "ENDIF
#>
#> $THETA
#> (0.001, 3) ; TVCL
#> (0.001, 34) ; TVV
#>
#> $OMEGA BLOCK(2)
#> 0.1 ; nCL
#> 0.1 0.1 ; nV
#>
#> $SIGMA
#> 0.03 ; PROP
#>
#> ; THETA PRIORS
#> $THETAP (3 FIX) (34 FIX)
#>
#> ; THETA (uniformative) PRIORs
#> $THETAPV BLOCK(2)
#> 10000 FIX
#> 0.0 10000
#>
#> $OMEGAP BLOCK(2)
#> 0.2 FIX
#> 0 0.2
#>
#> ; degrees of freedom to prior omega matrix - low dof = highly uninformative
#> $OMEGAPD (2 FIX)
#>
#> $EST METHOD=CHAIN FILE=..\run006chains.chn NSAMPLE=0 ISAMPLE=1 DF=20
#> $EST METHOD=BAYES INTER NBURN=4000 NITER=10000 PRINT=20 NOPRIOR=0 CTYPE=3 CITER=10 SEED=56664
#>
#> [[2]]
#> $PROB one cmpt simple mu modeled bayes
#> $SUB ADVAN1 TRANS2
#> $INPUT ID TIME DV CMT EVID AMT RATE
#> $DATA mdata/simple_nocovar_50id_6tp.csv IGNORE=@
#> $ABBR REPLACE THETA(CL, V) = THETA(1 to 2)
#> $ABBR REPLACE ETA(CL, V) = ETA(1 to 2)
#>
#> $THETAI
#> THETA(1:NTHETA)=LOG(THETAI(1:NTHETA))
#> THETAP(1:NTHETAP)=LOG(THETAPI(1:NTHETAP))
#> $THETAR
#> THETAR(1:NTHETA)=EXP(THETA(1:NTHETA))
#> THETAPR(1:NTHETAP)=EXP(THETAP(1:NTHETAP))
#>
#> $PRIOR NWPRI
#>
#> $PK
#> "USE NMBAYES_INT, ONLY: ITER_REPORT,BAYES_EXTRA_REQUEST,BAYES_EXTRA
#> ; Request extra information for Bayesian analysis.
#> ; An extra call will then be made for accepted samples
#> "BAYES_EXTRA_REQUEST=1
#>
#> MU_1 = THETA(CL)
#> MU_2 = THETA(V)
#> CL = EXP(MU_1 + ETA(CL))
#> V = EXP(MU_2 + ETA(V))
#> S1 = V
#>
#> "IF(BAYES_EXTRA==1 .AND. ITER_REPORT>=0 .AND. TIME==0.0) THEN
#> "WRITE(50,98) ITER_REPORT,ID,CL,V
#> "98 FORMAT(I12,1X,F14.0,4(1X,1PG12.5))
#> "ENDIF
#>
#> $ERROR
#> "USE NMBAYES_INT, ONLY: ITER_REPORT,BAYES_EXTRA_REQUEST,BAYES_EXTRA
#> "BAYES_EXTRA_REQUEST=1
#> IPRED=F
#> Y = IPRED*(1 + ERR(1))
#> "IF(BAYES_EXTRA==1 .AND. ITER_REPORT>=0 ) THEN
#> "WRITE(51,97) ITER_REPORT,ID,TIME,F
#> "97 FORMAT(I12,1X,F14.0,2(1X,1PG12.5))
#> "ENDIF
#>
#> $THETA
#> (0.001, 3) ; TVCL
#> (0.001, 34) ; TVV
#>
#> $OMEGA BLOCK(2)
#> 0.1 ; nCL
#> 0.1 0.1 ; nV
#>
#> $SIGMA
#> 0.03 ; PROP
#>
#> ; THETA PRIORS
#> $THETAP (3 FIX) (34 FIX)
#>
#> ; THETA (uniformative) PRIORs
#> $THETAPV BLOCK(2)
#> 10000 FIX
#> 0.0 10000
#>
#> $OMEGAP BLOCK(2)
#> 0.2 FIX
#> 0 0.2
#>
#> ; degrees of freedom to prior omega matrix - low dof = highly uninformative
#> $OMEGAPD (2 FIX)
#>
#> $EST METHOD=CHAIN FILE=..\run006chains.chn NSAMPLE=0 ISAMPLE=2 DF=20
#> $EST METHOD=BAYES INTER NBURN=4000 NITER=10000 PRINT=20 NOPRIOR=0 CTYPE=3 CITER=10 SEED=72923
#>
#> [[3]]
#> $PROB one cmpt simple mu modeled bayes
#> $SUB ADVAN1 TRANS2
#> $INPUT ID TIME DV CMT EVID AMT RATE
#> $DATA mdata/simple_nocovar_50id_6tp.csv IGNORE=@
#> $ABBR REPLACE THETA(CL, V) = THETA(1 to 2)
#> $ABBR REPLACE ETA(CL, V) = ETA(1 to 2)
#>
#> $THETAI
#> THETA(1:NTHETA)=LOG(THETAI(1:NTHETA))
#> THETAP(1:NTHETAP)=LOG(THETAPI(1:NTHETAP))
#> $THETAR
#> THETAR(1:NTHETA)=EXP(THETA(1:NTHETA))
#> THETAPR(1:NTHETAP)=EXP(THETAP(1:NTHETAP))
#>
#> $PRIOR NWPRI
#>
#> $PK
#> "USE NMBAYES_INT, ONLY: ITER_REPORT,BAYES_EXTRA_REQUEST,BAYES_EXTRA
#> ; Request extra information for Bayesian analysis.
#> ; An extra call will then be made for accepted samples
#> "BAYES_EXTRA_REQUEST=1
#>
#> MU_1 = THETA(CL)
#> MU_2 = THETA(V)
#> CL = EXP(MU_1 + ETA(CL))
#> V = EXP(MU_2 + ETA(V))
#> S1 = V
#>
#> "IF(BAYES_EXTRA==1 .AND. ITER_REPORT>=0 .AND. TIME==0.0) THEN
#> "WRITE(50,98) ITER_REPORT,ID,CL,V
#> "98 FORMAT(I12,1X,F14.0,4(1X,1PG12.5))
#> "ENDIF
#>
#> $ERROR
#> "USE NMBAYES_INT, ONLY: ITER_REPORT,BAYES_EXTRA_REQUEST,BAYES_EXTRA
#> "BAYES_EXTRA_REQUEST=1
#> IPRED=F
#> Y = IPRED*(1 + ERR(1))
#> "IF(BAYES_EXTRA==1 .AND. ITER_REPORT>=0 ) THEN
#> "WRITE(51,97) ITER_REPORT,ID,TIME,F
#> "97 FORMAT(I12,1X,F14.0,2(1X,1PG12.5))
#> "ENDIF
#>
#> $THETA
#> (0.001, 3) ; TVCL
#> (0.001, 34) ; TVV
#>
#> $OMEGA BLOCK(2)
#> 0.1 ; nCL
#> 0.1 0.1 ; nV
#>
#> $SIGMA
#> 0.03 ; PROP
#>
#> ; THETA PRIORS
#> $THETAP (3 FIX) (34 FIX)
#>
#> ; THETA (uniformative) PRIORs
#> $THETAPV BLOCK(2)
#> 10000 FIX
#> 0.0 10000
#>
#> $OMEGAP BLOCK(2)
#> 0.2 FIX
#> 0 0.2
#>
#> ; degrees of freedom to prior omega matrix - low dof = highly uninformative
#> $OMEGAPD (2 FIX)
#>
#> $EST METHOD=CHAIN FILE=..\run006chains.chn NSAMPLE=0 ISAMPLE=3 DF=20
#> $EST METHOD=BAYES INTER NBURN=4000 NITER=10000 PRINT=20 NOPRIOR=0 CTYPE=3 CITER=10 SEED=91610
#>
#> [[4]]
#> $PROB one cmpt simple mu modeled bayes
#> $SUB ADVAN1 TRANS2
#> $INPUT ID TIME DV CMT EVID AMT RATE
#> $DATA mdata/simple_nocovar_50id_6tp.csv IGNORE=@
#> $ABBR REPLACE THETA(CL, V) = THETA(1 to 2)
#> $ABBR REPLACE ETA(CL, V) = ETA(1 to 2)
#>
#> $THETAI
#> THETA(1:NTHETA)=LOG(THETAI(1:NTHETA))
#> THETAP(1:NTHETAP)=LOG(THETAPI(1:NTHETAP))
#> $THETAR
#> THETAR(1:NTHETA)=EXP(THETA(1:NTHETA))
#> THETAPR(1:NTHETAP)=EXP(THETAP(1:NTHETAP))
#>
#> $PRIOR NWPRI
#>
#> $PK
#> "USE NMBAYES_INT, ONLY: ITER_REPORT,BAYES_EXTRA_REQUEST,BAYES_EXTRA
#> ; Request extra information for Bayesian analysis.
#> ; An extra call will then be made for accepted samples
#> "BAYES_EXTRA_REQUEST=1
#>
#> MU_1 = THETA(CL)
#> MU_2 = THETA(V)
#> CL = EXP(MU_1 + ETA(CL))
#> V = EXP(MU_2 + ETA(V))
#> S1 = V
#>
#> "IF(BAYES_EXTRA==1 .AND. ITER_REPORT>=0 .AND. TIME==0.0) THEN
#> "WRITE(50,98) ITER_REPORT,ID,CL,V
#> "98 FORMAT(I12,1X,F14.0,4(1X,1PG12.5))
#> "ENDIF
#>
#> $ERROR
#> "USE NMBAYES_INT, ONLY: ITER_REPORT,BAYES_EXTRA_REQUEST,BAYES_EXTRA
#> "BAYES_EXTRA_REQUEST=1
#> IPRED=F
#> Y = IPRED*(1 + ERR(1))
#> "IF(BAYES_EXTRA==1 .AND. ITER_REPORT>=0 ) THEN
#> "WRITE(51,97) ITER_REPORT,ID,TIME,F
#> "97 FORMAT(I12,1X,F14.0,2(1X,1PG12.5))
#> "ENDIF
#>
#> $THETA
#> (0.001, 3) ; TVCL
#> (0.001, 34) ; TVV
#>
#> $OMEGA BLOCK(2)
#> 0.1 ; nCL
#> 0.1 0.1 ; nV
#>
#> $SIGMA
#> 0.03 ; PROP
#>
#> ; THETA PRIORS
#> $THETAP (3 FIX) (34 FIX)
#>
#> ; THETA (uniformative) PRIORs
#> $THETAPV BLOCK(2)
#> 10000 FIX
#> 0.0 10000
#>
#> $OMEGAP BLOCK(2)
#> 0.2 FIX
#> 0 0.2
#>
#> ; degrees of freedom to prior omega matrix - low dof = highly uninformative
#> $OMEGAPD (2 FIX)
#>
#> $EST METHOD=CHAIN FILE=..\run006chains.chn NSAMPLE=0 ISAMPLE=4 DF=20
#> $EST METHOD=BAYES INTER NBURN=4000 NITER=10000 PRINT=20 NOPRIOR=0 CTYPE=3 CITER=10 SEED=3940r6c1 <- fread("../modeling/run006c1.ext", skip = 1) %>% mutate(chain = 1)
r6c2 <- fread("../modeling/run006c2.ext", skip = 1) %>% mutate(chain = 2)
r6c3 <- fread("../modeling/run006c3.ext", skip = 1) %>% mutate(chain = 3)
r6c4 <- fread("../modeling/run006c4.ext", skip = 1) %>% mutate(chain = 4)r6_chains <- bind_rows(r6c1, r6c2, r6c3) %>%
filter(ITERATION > 0) %>%
rename(CL = THETA1,
V = THETA2,
EPS = `SIGMA(1,1)`,
nCL = `OMEGA(1,1)`,
nV = `OMEGA(2,2)`,
nCL_nV = `OMEGA(2,1)`)kable(head(r6_chains))| ITERATION | CL | V | EPS | nCL | nCL_nV | nV | MCMCOBJ | chain |
|---|---|---|---|---|---|---|---|---|
| 1 | 3.08 | 35.7 | 0.044 | 0.121 | 0.025 | 0.054 | 463 | 1 |
| 2 | 2.96 | 35.4 | 0.042 | 0.140 | 0.010 | 0.044 | 465 | 1 |
| 3 | 3.41 | 32.8 | 0.044 | 0.116 | 0.030 | 0.044 | 484 | 1 |
| 4 | 3.33 | 35.4 | 0.043 | 0.150 | 0.026 | 0.049 | 515 | 1 |
| 5 | 3.35 | 35.2 | 0.053 | 0.162 | 0.006 | 0.044 | 495 | 1 |
| 6 | 3.35 | 35.2 | 0.049 | 0.099 | 0.009 | 0.037 | 467 | 1 |
thin_by <- function(df, .mod) {
df %>% filter(ITERATION %% .mod == 0)
}r6_chains %>% select(ITERATION:nV, chain) %>%
thin_by(10) %>%
gather(param, value, CL:nV) %>%
ggplot(aes(x = ITERATION, y = value, color = factor(chain)), alpha = 0.6) +
geom_line() + facet_wrap(~param, ncol = 2, scales = "free") +
theme_bw() +
base_theme() +
scale_color_discrete(name = "Chain") +
theme(legend.position = "bottom") 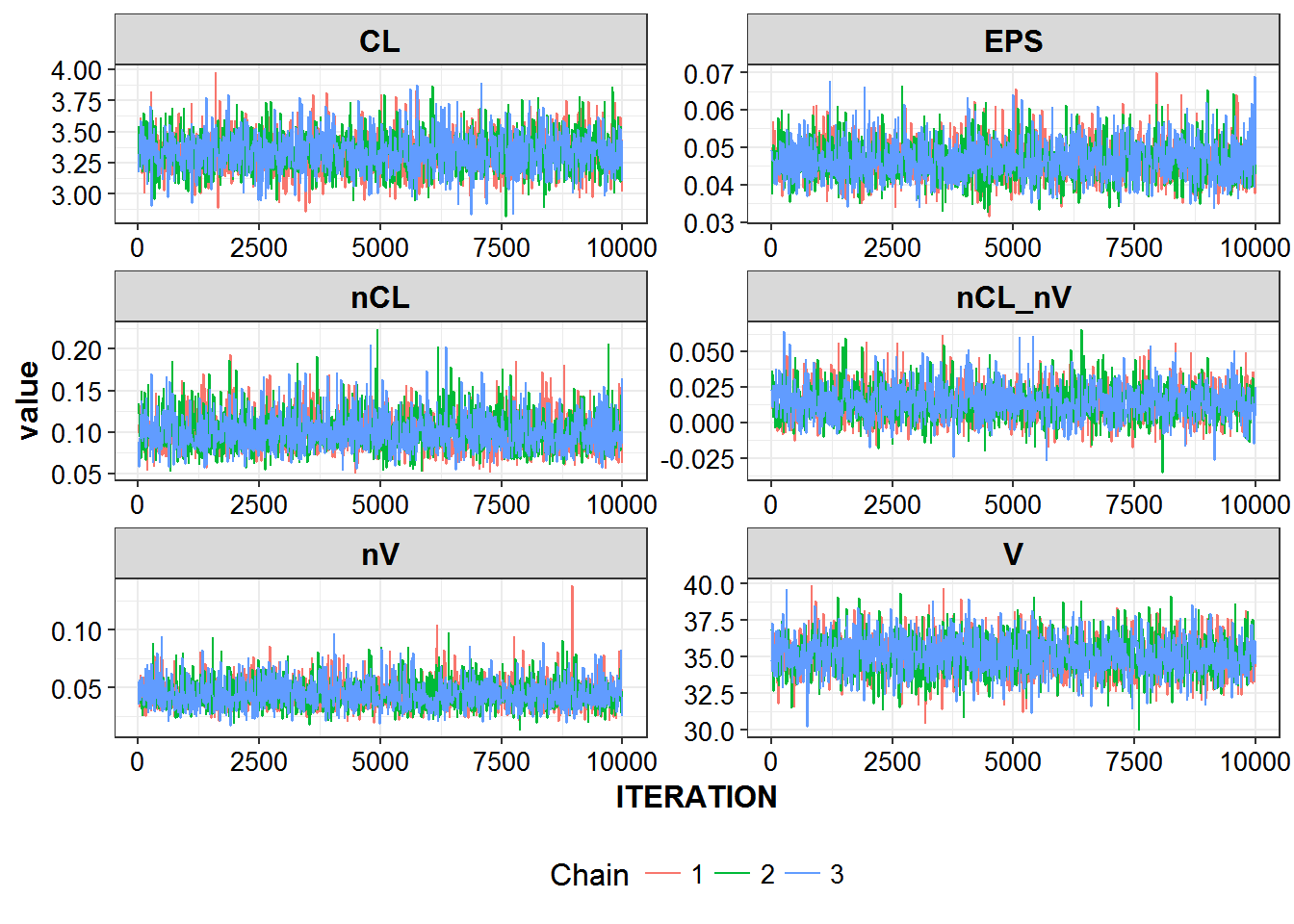
session_details <- devtools::session_info()
session_details$platform
#> setting value
#> version R version 3.3.2 (2016-10-31)
#> system x86_64, mingw32
#> ui RTerm
#> language (EN)
#> collate English_United States.1252
#> tz America/New_York
#> date 2016-12-13
knitr::kable(session_details$packages)| package | * | version | date | source |
|---|---|---|---|---|
| backports | 1.0.4 | 2016-10-24 | CRAN (R 3.3.2) | |
| bookdown | 0.2 | 2016-11-12 | CRAN (R 3.3.2) | |
| devtools | 1.12.0 | 2016-06-24 | CRAN (R 3.3.2) | |
| digest | 0.6.10 | 2016-08-02 | CRAN (R 3.3.2) | |
| evaluate | 0.10 | 2016-10-11 | CRAN (R 3.3.2) | |
| htmltools | 0.3.5 | 2016-03-21 | CRAN (R 3.3.2) | |
| httpuv | 1.3.3 | 2015-08-04 | CRAN (R 3.3.2) | |
| knitr | 1.15 | 2016-11-09 | CRAN (R 3.3.2) | |
| magrittr | 1.5 | 2014-11-22 | CRAN (R 3.3.2) | |
| memoise | 1.0.0 | 2016-01-29 | CRAN (R 3.3.2) | |
| mime | 0.5 | 2016-07-07 | CRAN (R 3.3.2) | |
| miniUI | 0.1.1 | 2016-01-15 | CRAN (R 3.3.2) | |
| R6 | 2.2.0 | 2016-10-05 | CRAN (R 3.3.2) | |
| Rcpp | 0.12.8 | 2016-11-17 | CRAN (R 3.3.2) | |
| rmarkdown | 1.2 | 2016-11-21 | CRAN (R 3.3.2) | |
| rprojroot | 1.1 | 2016-10-29 | CRAN (R 3.3.2) | |
| shiny | 0.14.2 | 2016-11-01 | CRAN (R 3.3.2) | |
| stringi | 1.1.2 | 2016-10-01 | CRAN (R 3.3.2) | |
| stringr | 1.1.0 | 2016-08-19 | CRAN (R 3.3.2) | |
| withr | 1.0.2 | 2016-06-20 | CRAN (R 3.3.2) | |
| xtable | 1.8-2 | 2016-02-05 | CRAN (R 3.3.2) | |
| yaml | 2.1.13 | 2014-06-12 | CRAN (R 3.3.2) |