5 Dataset Creation
library(knitr)
library(PKPDmisc)
library(tidyverse)
library(mrgsolve)
library(infuser)source("../scripts/model_details.R")models <- source("../models/models.R")$value
#> model dir set to C:/Users/devin/Documents/Repos/simplest_bayes/models
#> cache location set to C:\Users\devin\Documents\Repos\simplest_bayes\models\.modelcache
#> Loading model from cache.5.1 Generate data for mrgsolve
start with a baseline of having 50 individuals worth of data, can scale to different amounts of individuals later
NIDS <- 56demogs <- data_frame(ID = 1:NIDS)for_dosing <- demogs %>%
mutate(
CMT = 1,
EVID = 1,
TIME = 0,
AMT = 1000,
RATE = 1000,
ADDL = 1,
II = 12,
OBSNUM = 0
) one_cmt_iv <- models$use("one_cmt_iv")5.2 Model Details
mrgsolve::see(one_cmt_iv)
#>
#> Model file: one_cmt_iv.cpp
#> [PARAM] @annotated
#> CL : 3 : Clearance (L/hr)
#> V : 35 : Volume (L)
#>
#>
#> [CMT] @annotated
#> CENT : Central compartment (mg)
#>
#> [PKMODEL]
#> ncmt=1, trans=11
#>
#> [MAIN]
#> double CLi = CL*exp(nCL);
#> double Vi = V*exp(nV);
#>
#>
#> [OMEGA] @annotated @correlation @block
#> nCL : 0.1 : Random effect on CL
#> nV : 0.4 0.04 : Random effect on V
#>
#> [SIGMA] @annotated
#> PROP : 0.04 : Proportional error
#> // so don't get into issues with estimating via multiplicative error only
#> ADD : 0.1 : Additive residual error
#>
#> [TABLE]
#> double IPRED = CENT/Vi;
#> double DV = CENT/Vi*(1+PROP) + ADD;
#>
#> [CAPTURE] @annotated
#> DV : plasma concentration (mg/L)
#> IPRED : Individual predicted plasma concentration (mg/L)
#> CLi : Individual Clearance (L/hr)
#> Vi : Individual Volume (L)one_cmt_iv %>%
model_details %>%
filter(block != "CAPTURE") %>%
kable()| block | name | descr | unit | options | value |
|---|---|---|---|---|---|
| PARAM | CL | Clearance | L/hr | . | 3.00 |
| PARAM | V | Volume | L | . | 35.00 |
| CMT | CENT | Central compartment | mg | . | 0.00 |
| OMEGA | nCL | Random effect on CL | . | . | 0.10 |
| OMEGA | nV | Random effect on V | . | . | 0.04 |
| SIGMA | PROP | Proportional error | . | . | 0.04 |
| SIGMA | ADD | Additive residual error | . | . | 0.10 |
simulated_data <- one_cmt_iv %>%
data_set(for_dosing) %>%
mrgsim(end = 36, delta = 0.25) %>% as_data_frame5.3 Distribution of peak and trough values
- ‘peak’ defined as 1 hr post infusion and trough 1 hour prior to when next dose would begin
simulated_data %>%
filter(TIME %in% c(2, 11)) %>%
mutate(DV = ifelse(DV < 0, 0, DV)) %>%
select(ID, DV) %>%
group_by(ID) %>%
summarize_all(funs(min, max)) %>%
gather(sample, value, -ID) %>%
ggplot(aes(x = value)) +
geom_density() +
facet_wrap(~sample, scales = "free") +
theme_bw() +
base_theme()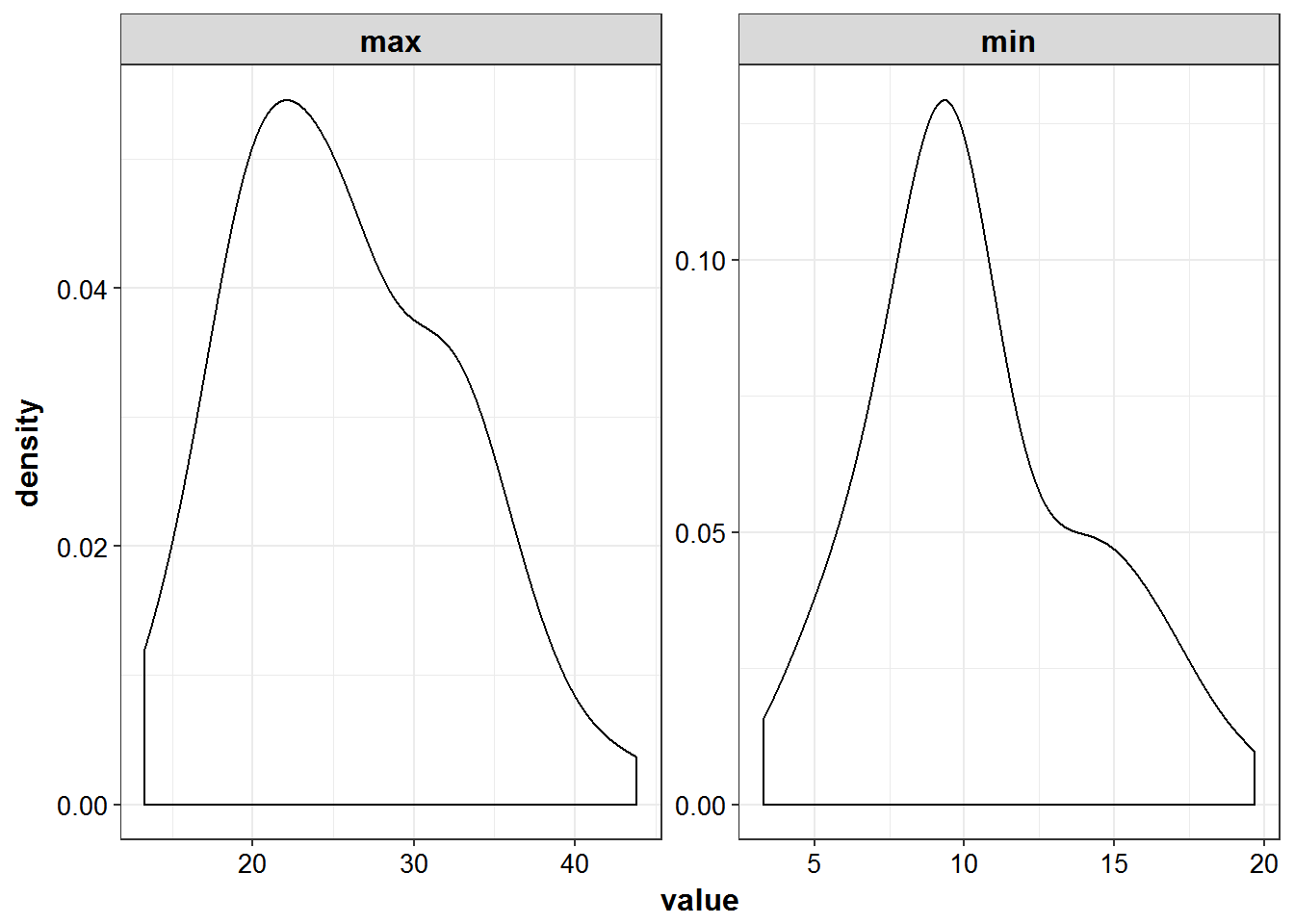
5.4 Predicted Profiles
simulated_data %>%
filter(IPRED > 0.1) %>%
ggplot(aes(x = TIME, y = IPRED, group = ID)) +
geom_line(size = 1.05, alpha = 0.8) + theme_bw() +
base_theme() + scale_y_log10(breaks = c(0.1, 1, 10, 20, 30, 40))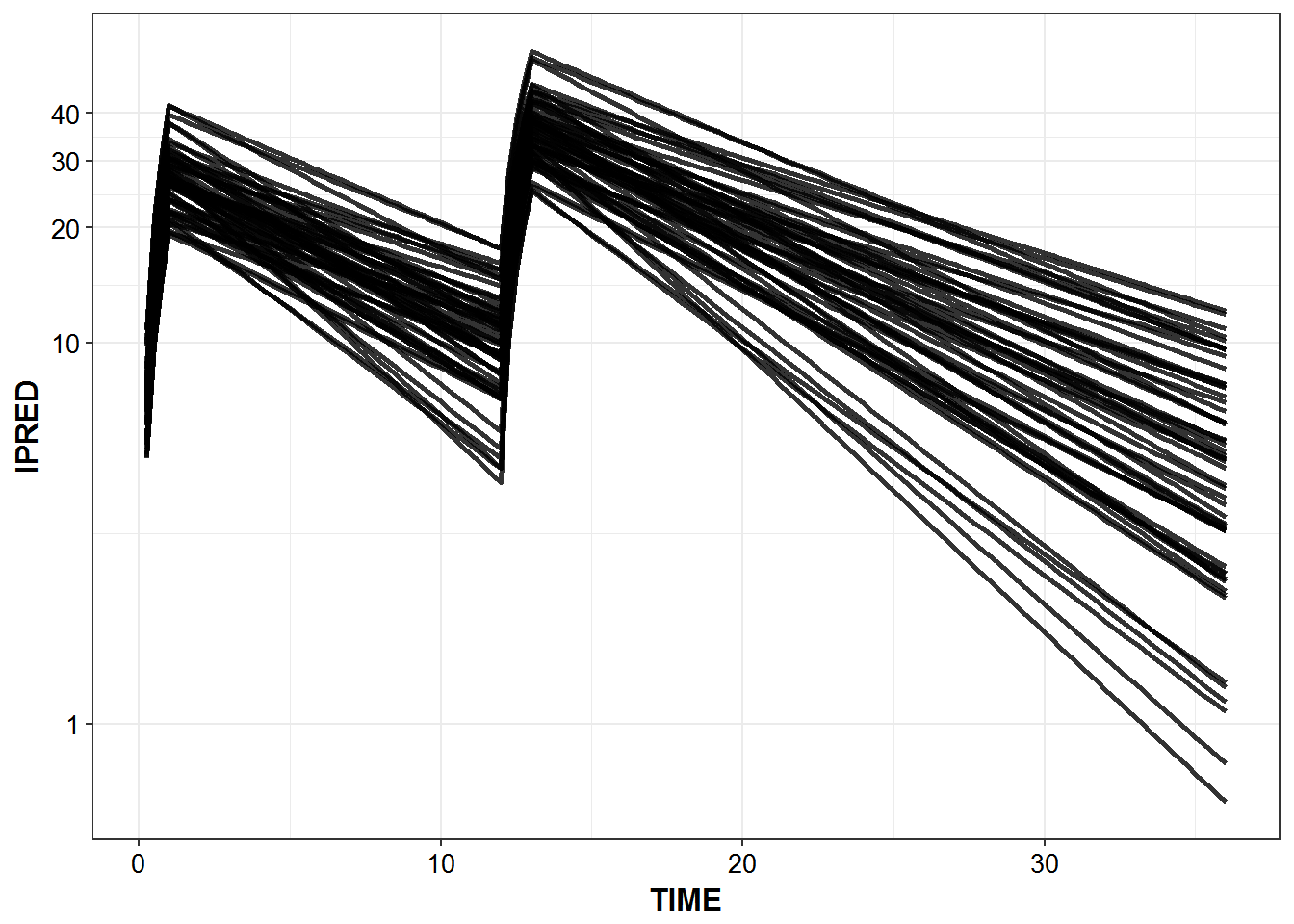
5.5 Real world sampling and LLOQ
sample_times_rich <- c(2, 6, 11, 14, 18, 23)
LLOQ <- 0.1
sampled_data_rich2d <- simulated_data %>%
filter(TIME %in% sample_times_rich, DV > LLOQ) %>%
group_by(ID) %>% mutate(OBSNUM = dplyr::row_number(ID))
sparser_scenarios <- list(
"s2d" = c(1, 3, 4, 6),
"s1dpt" = c(1, 3, 6),
"r1d" = c(1, 2, 3),
"s2trough" = c(3, 6),
"s1d" = c(1, 3),
"tr" = c(3)
)
sparser_scenario_data <- map(names(sparser_scenarios), function(scenario) {
res <- sampled_data_rich2d %>%
filter(OBSNUM %in% sparser_scenarios[[scenario]])
return(setNames(res, scenario))
})5.6 IPRED and DV vs TIME for all individuals at sampled times
list_plots_rich <- simulated_data %>%
select(-DV) %>%
left_join(sampled_data_rich2d %>% select(ID, TIME, DV)) %>%
mutate(PNUM = ids_per_plot(ID)) %>%
split(.$PNUM) %>%
map(~
ggplot(., aes(x = TIME, y = IPRED, group = ID)) +
geom_point(aes(y = DV), color = "blue") +
geom_line(size = 1.05, alpha = 0.8) + theme_bw() +
base_theme() + facet_wrap(~ID) +
scale_y_log10(breaks = c(0.1, 1, 5, 10, 20, 30, 40))
)
#> Joining, by = c("ID", "TIME")
print_plots(list_plots_rich)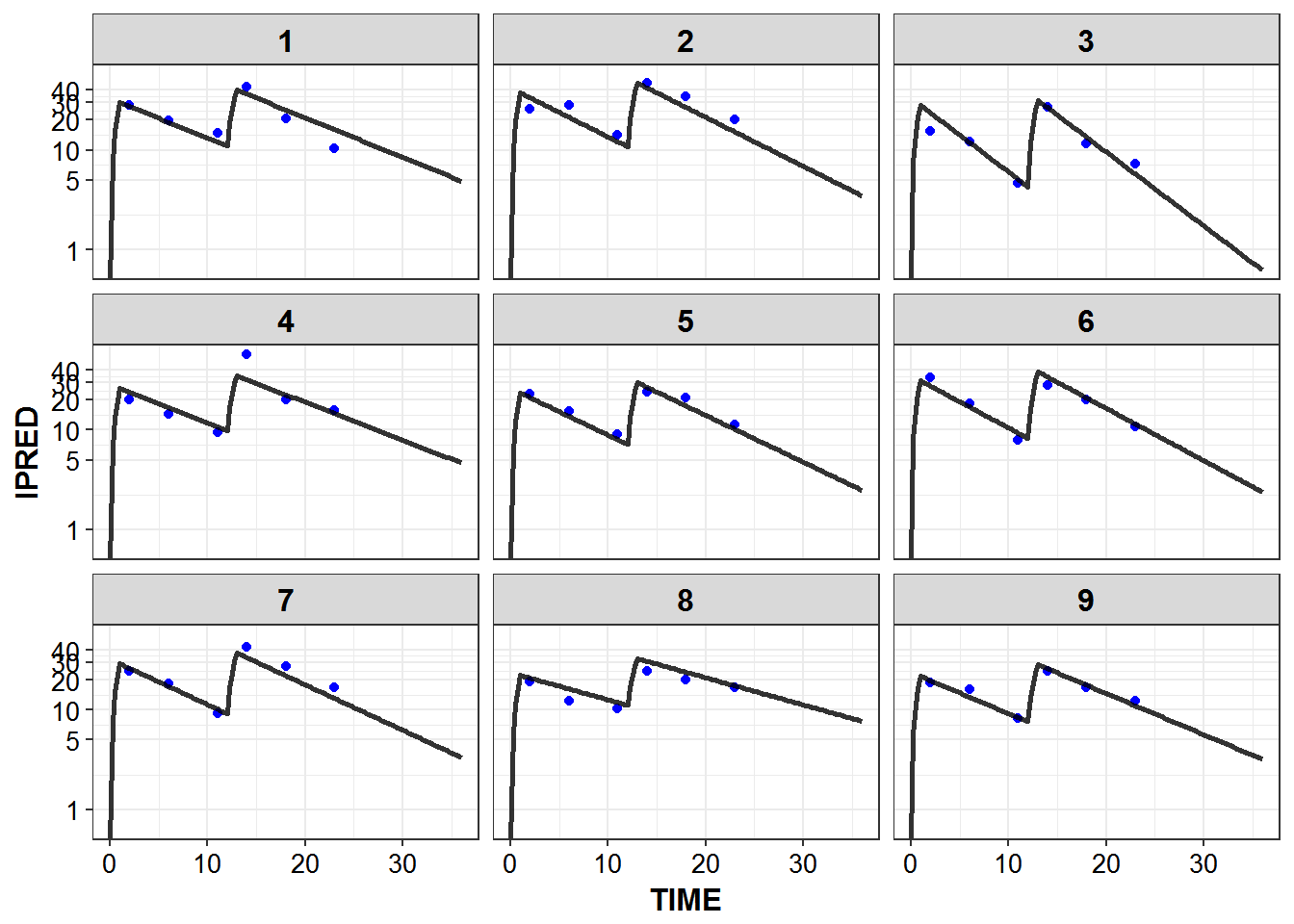
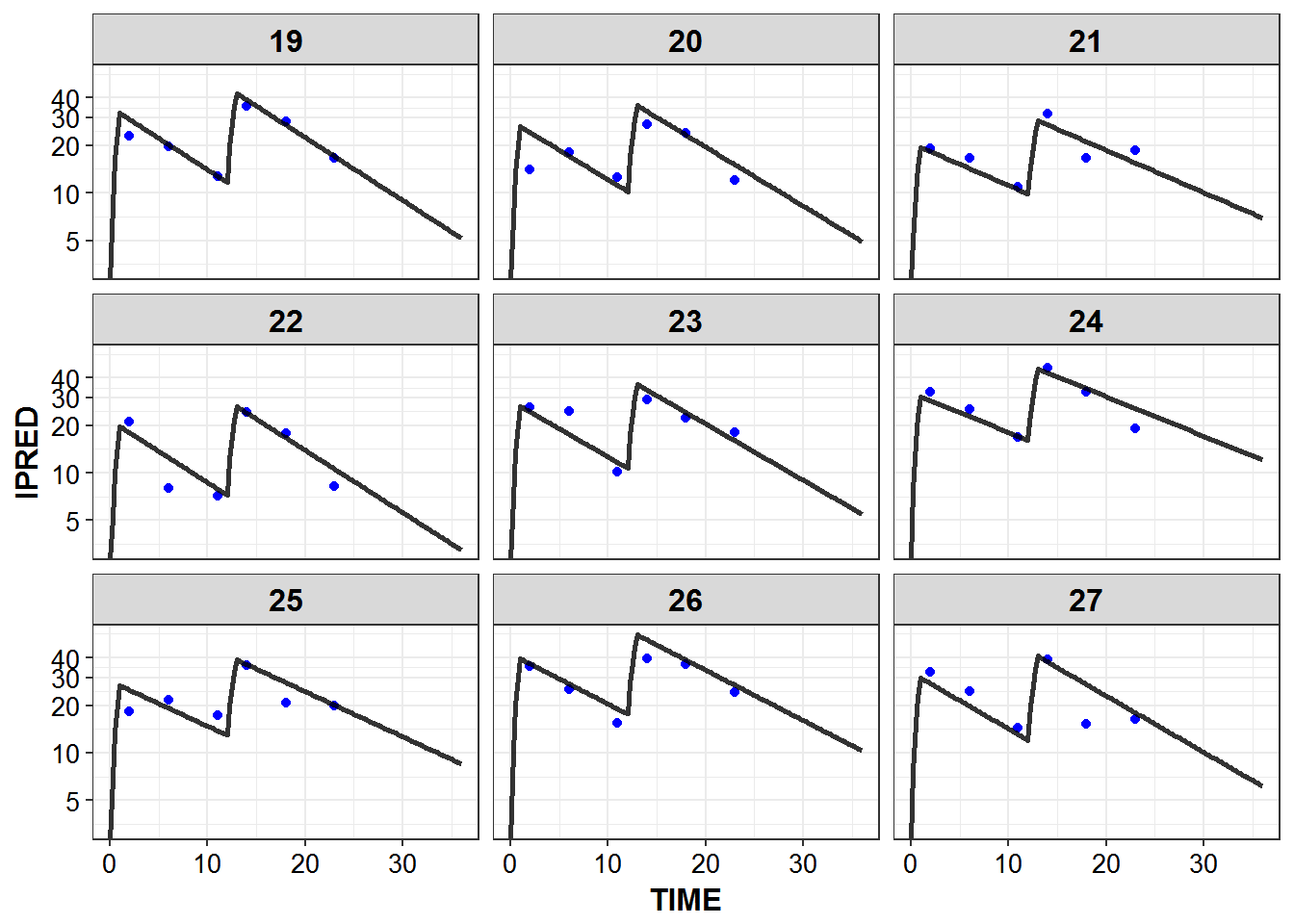
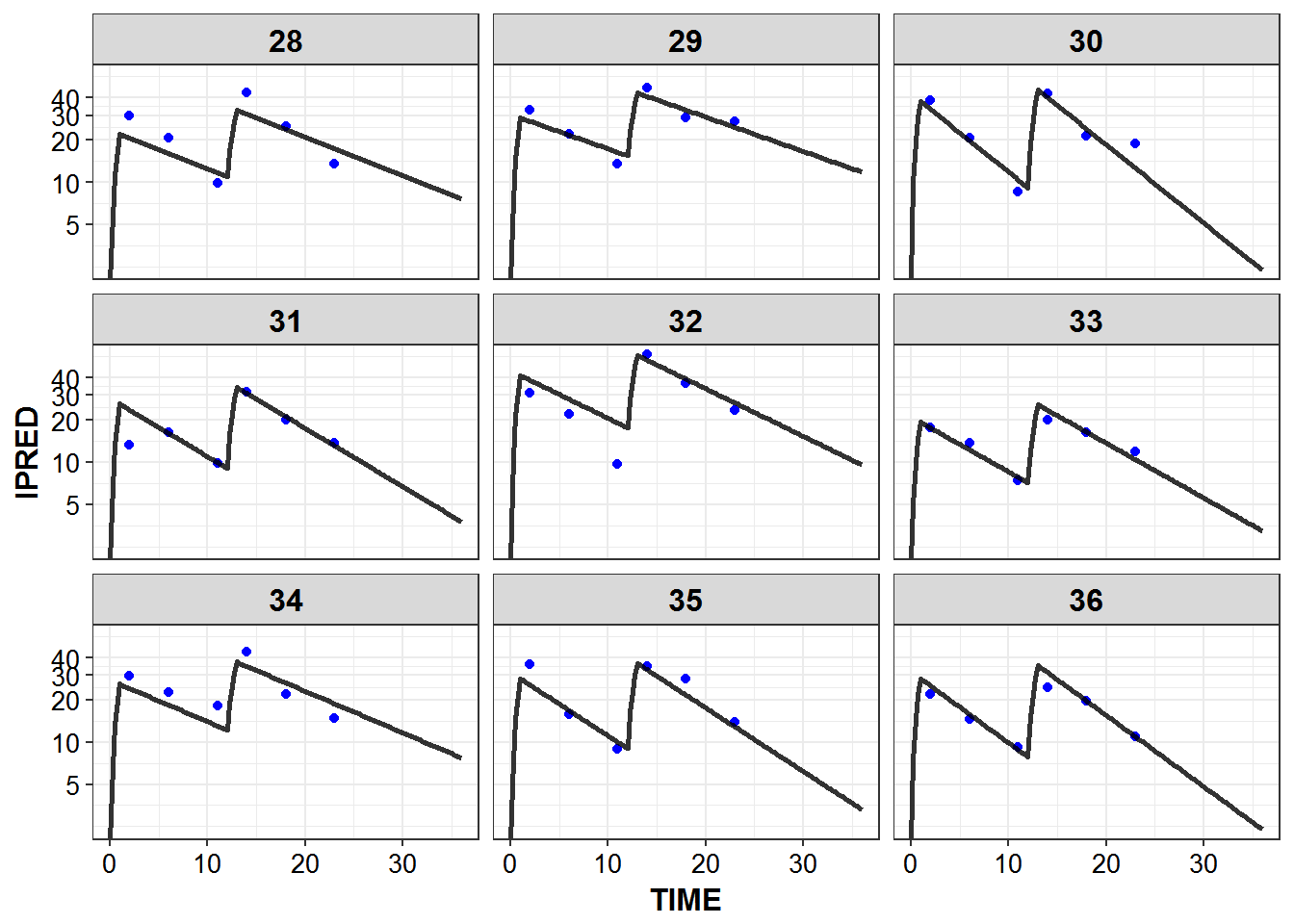
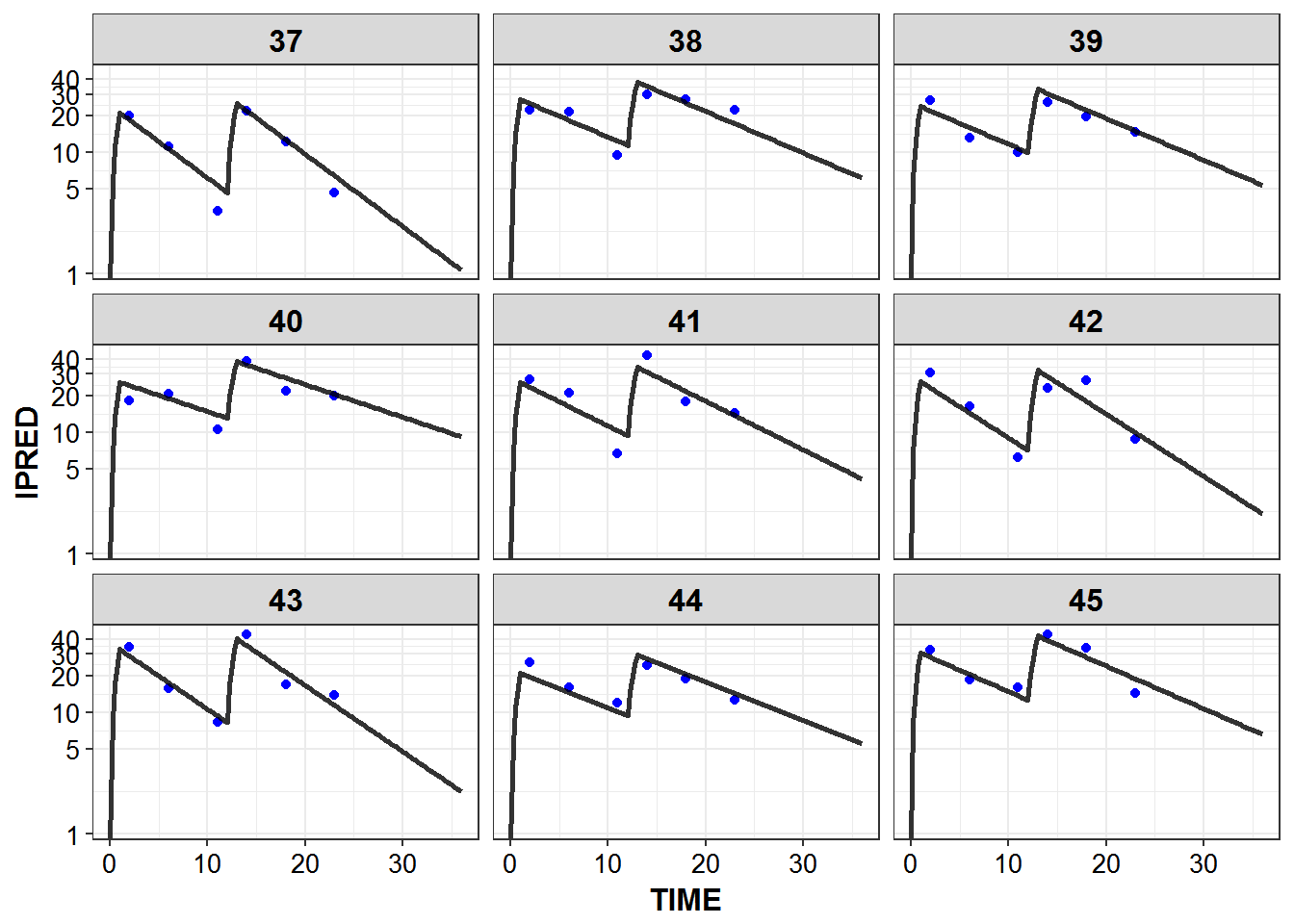
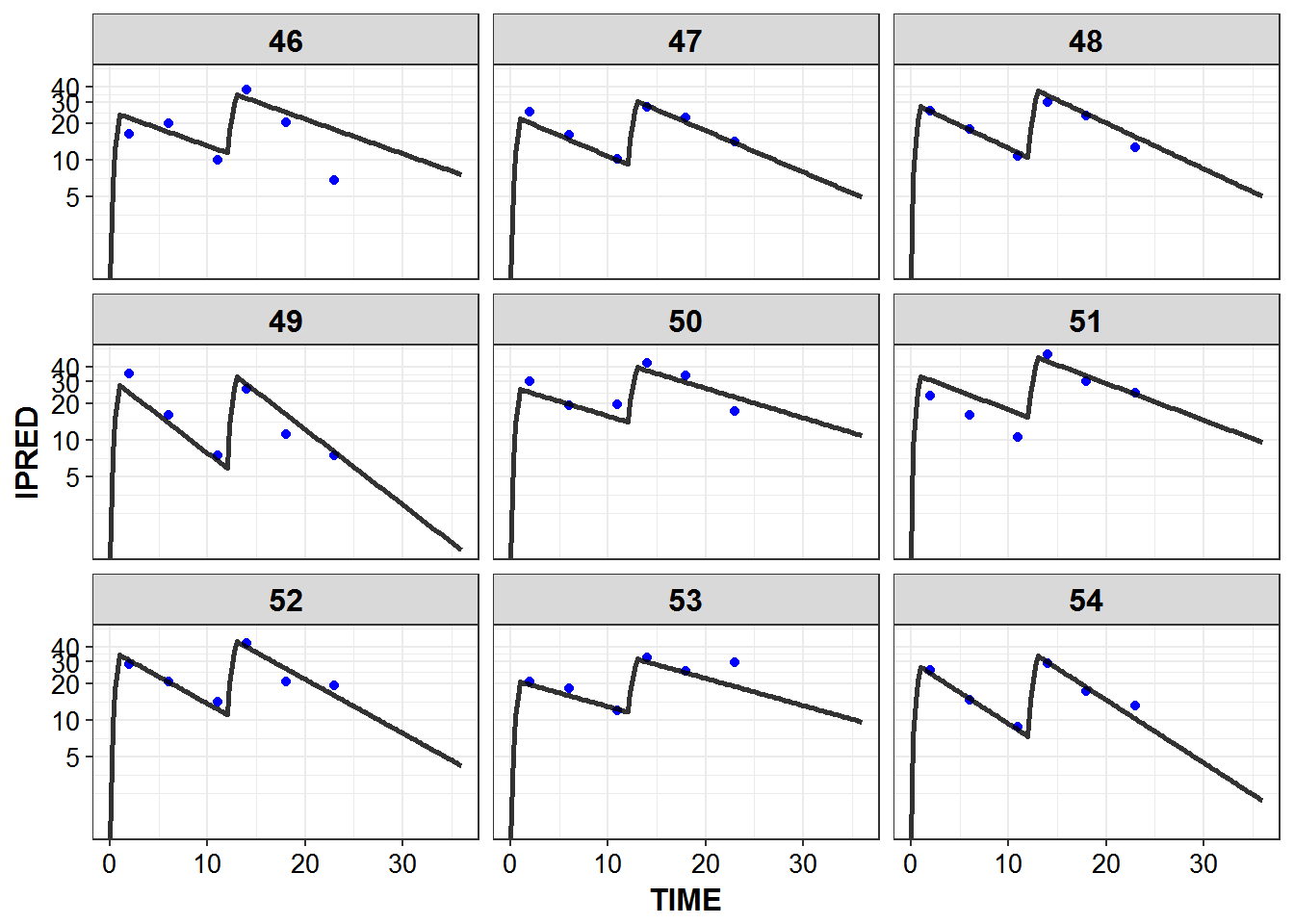
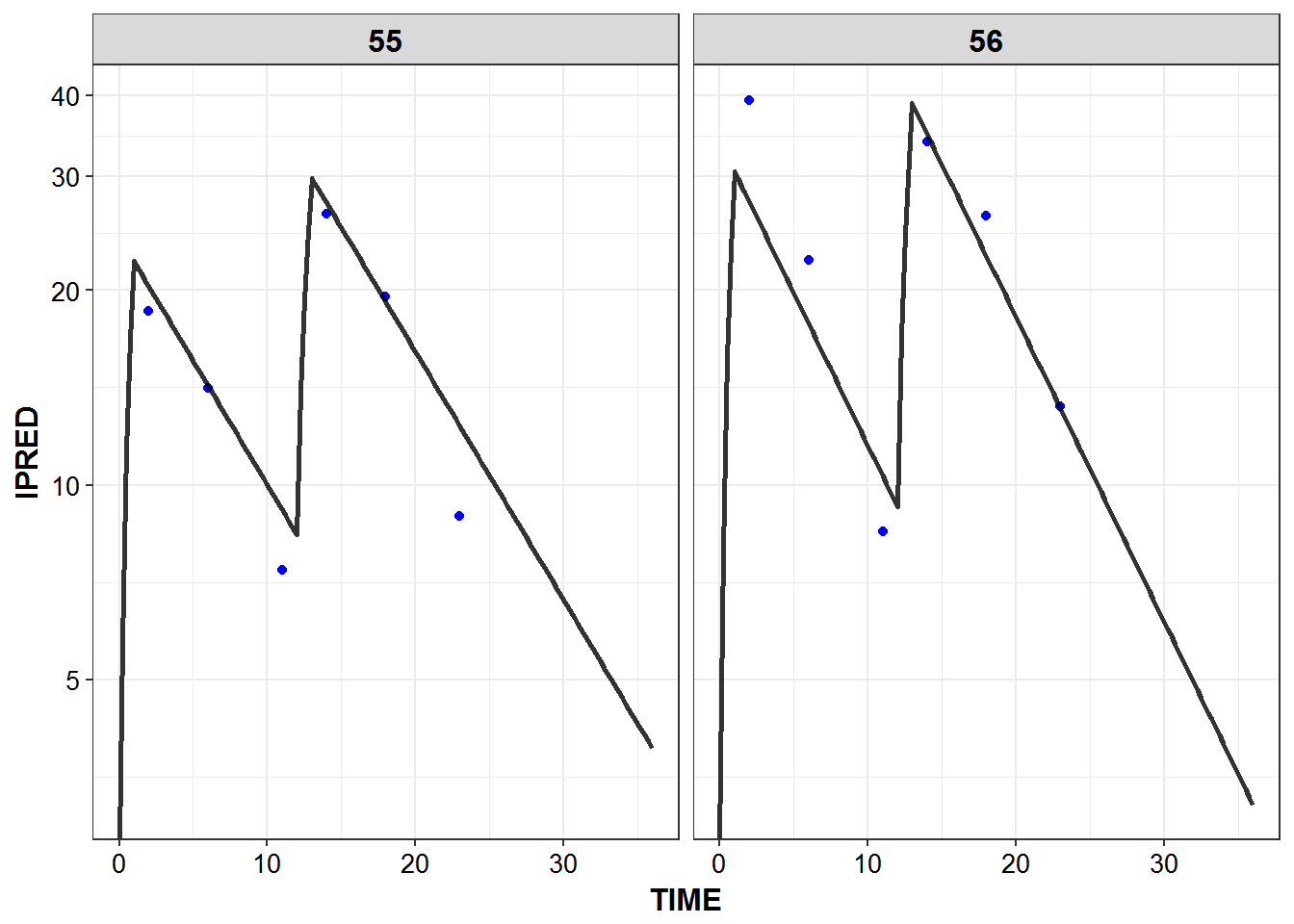
#> [[1]]
#> NULL
#>
#> [[2]]
#> NULL
#>
#> [[3]]
#> NULL
#>
#> [[4]]
#> NULL
#>
#> [[5]]
#> NULL
#>
#> [[6]]
#> NULL
#>
#> [[7]]
#> NULL5.7 Prepare for nonmem
nm_dat_rich <- sampled_data_rich2d %>% select(ID, TIME, DV, OBSNUM) %>%
mutate(
CMT = 1,
EVID = 0
) %>%
bind_rows(for_dosing) %>%
arrange(ID, TIME, desc(EVID))*rich data
kable(head(nm_dat_rich, n = 14))| ID | TIME | DV | OBSNUM | CMT | EVID | AMT | RATE | ADDL | II |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | NA | 0 | 1 | 1 | 1000 | 1000 | 1 | 12 |
| 1 | 2 | 28.0 | 1 | 1 | 0 | NA | NA | NA | NA |
| 1 | 6 | 19.7 | 2 | 1 | 0 | NA | NA | NA | NA |
| 1 | 11 | 14.9 | 3 | 1 | 0 | NA | NA | NA | NA |
| 1 | 14 | 43.1 | 4 | 1 | 0 | NA | NA | NA | NA |
| 1 | 18 | 20.7 | 5 | 1 | 0 | NA | NA | NA | NA |
| 1 | 23 | 10.3 | 6 | 1 | 0 | NA | NA | NA | NA |
| 2 | 0 | NA | 0 | 1 | 1 | 1000 | 1000 | 1 | 12 |
| 2 | 2 | 26.1 | 1 | 1 | 0 | NA | NA | NA | NA |
| 2 | 6 | 28.3 | 2 | 1 | 0 | NA | NA | NA | NA |
| 2 | 11 | 14.1 | 3 | 1 | 0 | NA | NA | NA | NA |
| 2 | 14 | 46.5 | 4 | 1 | 0 | NA | NA | NA | NA |
| 2 | 18 | 34.4 | 5 | 1 | 0 | NA | NA | NA | NA |
| 2 | 23 | 20.1 | 6 | 1 | 0 | NA | NA | NA | NA |
5.8 Create chains
modt <- read_file("../modeling/run007c.modt")
BASE_MODEL_NUM <- 7
scenario_df <- as_data_frame(
expand.grid(chain = 1:4, scenario = names(sparser_scenarios))
) %>% arrange(scenario) %>%
mutate(scenario_num = BASE_MODEL_NUM + as.numeric(as.factor(scenario)))
kable(scenario_df)| chain | scenario | scenario_num |
|---|---|---|
| 1 | s2d | 8 |
| 2 | s2d | 8 |
| 3 | s2d | 8 |
| 4 | s2d | 8 |
| 1 | s1dpt | 9 |
| 2 | s1dpt | 9 |
| 3 | s1dpt | 9 |
| 4 | s1dpt | 9 |
| 1 | r1d | 10 |
| 2 | r1d | 10 |
| 3 | r1d | 10 |
| 4 | r1d | 10 |
| 1 | s2trough | 11 |
| 2 | s2trough | 11 |
| 3 | s2trough | 11 |
| 4 | s2trough | 11 |
| 1 | s1d | 12 |
| 2 | s1d | 12 |
| 3 | s1d | 12 |
| 4 | s1d | 12 |
| 1 | tr | 13 |
| 2 | tr | 13 |
| 3 | tr | 13 |
| 4 | tr | 13 |
Want to inject in the chain number and scenario name, as well as subset the data relevant to the scenario. The ignore statements control which OBSNUM will be retained for estimation
by_row(scenario_df, function(row) {
set.seed(1234567)
ignore_obs <- setdiff(1:length(sample_times_rich), sparser_scenarios[[row$scenario]])
ignore <- paste0("IGNORE=(OBSNUM.EQN.", ignore_obs, ")", collapse = " ")
write_file(
infuse(modt,
chain_number = row$chain,
seed = round(runif(1, 1000, 100000), 0),
ignore = ignore,
scenario = row$scenario),
file.path("..", "modeling", paste0("run", pad_left(row$scenario_num, 3), "c", row$chain, ".mod")))
})
#> # A tibble: 24 × 4
#> chain scenario scenario_num .out
#> <int> <fctr> <dbl> <list>
#> 1 1 s2d 8 <S3: character>
#> 2 2 s2d 8 <S3: character>
#> 3 3 s2d 8 <S3: character>
#> 4 4 s2d 8 <S3: character>
#> 5 1 s1dpt 9 <S3: character>
#> 6 2 s1dpt 9 <S3: character>
#> # ... with 18 more rows5.9 output data
write_nonmem(nm_dat_rich, "../modeling/mdata/simple_nocovar_56id_6tp_md.csv")session_details <- devtools::session_info()
session_details$platform
#> setting value
#> version R version 3.3.2 (2016-10-31)
#> system x86_64, mingw32
#> ui RTerm
#> language (EN)
#> collate English_United States.1252
#> tz America/New_York
#> date 2016-12-13
knitr::kable(session_details$packages)| package | * | version | date | source |
|---|---|---|---|---|
| backports | 1.0.4 | 2016-10-24 | CRAN (R 3.3.2) | |
| bookdown | 0.2 | 2016-11-12 | CRAN (R 3.3.2) | |
| devtools | 1.12.0 | 2016-06-24 | CRAN (R 3.3.2) | |
| digest | 0.6.10 | 2016-08-02 | CRAN (R 3.3.2) | |
| evaluate | 0.10 | 2016-10-11 | CRAN (R 3.3.2) | |
| htmltools | 0.3.5 | 2016-03-21 | CRAN (R 3.3.2) | |
| httpuv | 1.3.3 | 2015-08-04 | CRAN (R 3.3.2) | |
| knitr | 1.15 | 2016-11-09 | CRAN (R 3.3.2) | |
| magrittr | 1.5 | 2014-11-22 | CRAN (R 3.3.2) | |
| memoise | 1.0.0 | 2016-01-29 | CRAN (R 3.3.2) | |
| mime | 0.5 | 2016-07-07 | CRAN (R 3.3.2) | |
| miniUI | 0.1.1 | 2016-01-15 | CRAN (R 3.3.2) | |
| R6 | 2.2.0 | 2016-10-05 | CRAN (R 3.3.2) | |
| Rcpp | 0.12.8 | 2016-11-17 | CRAN (R 3.3.2) | |
| rmarkdown | 1.2 | 2016-11-21 | CRAN (R 3.3.2) | |
| rprojroot | 1.1 | 2016-10-29 | CRAN (R 3.3.2) | |
| shiny | 0.14.2 | 2016-11-01 | CRAN (R 3.3.2) | |
| stringi | 1.1.2 | 2016-10-01 | CRAN (R 3.3.2) | |
| stringr | 1.1.0 | 2016-08-19 | CRAN (R 3.3.2) | |
| withr | 1.0.2 | 2016-06-20 | CRAN (R 3.3.2) | |
| xtable | 1.8-2 | 2016-02-05 | CRAN (R 3.3.2) | |
| yaml | 2.1.13 | 2014-06-12 | CRAN (R 3.3.2) |