1 Dataset Creation
library(knitr)
library(PKPDmisc)
library(tidyverse)
library(mrgsolve) source("../scripts/model_details.R")models <- source("../models/models.R")$value
#> model dir set to C:/Users/devin/Documents/Repos/simplest_bayes/models
#> cache location set to C:\Users\devin\Documents\Repos\simplest_bayes\models\.modelcache
#> Loading model from cache.1.1 Generate data for mrgsolve
start with a baseline of having 50 individuals worth of data, can scale to different amounts of individuals later
NIDS <- 50demogs <- data_frame(ID = 1:NIDS)for_dosing <- demogs %>%
mutate(
CMT = 1,
EVID = 1,
TIME = 0,
AMT = 1000,
RATE = 1000
) one_cmt_iv <- models$use("one_cmt_iv")1.2 Model Details
mrgsolve::see(one_cmt_iv)
#>
#> Model file: one_cmt_iv.cpp
#> [PARAM] @annotated
#> CL : 3 : Clearance (L/hr)
#> V : 35 : Volume (L)
#>
#>
#> [CMT] @annotated
#> CENT : Central compartment (mg)
#>
#> [PKMODEL]
#> ncmt=1, trans=11
#>
#> [MAIN]
#> double CLi = CL*exp(nCL);
#> double Vi = V*exp(nV);
#>
#>
#> [OMEGA] @annotated @correlation @block
#> nCL : 0.1 : Random effect on CL
#> nV : 0.4 0.04 : Random effect on V
#>
#> [SIGMA] @annotated
#> PROP : 0.04 : Proportional error
#> // so don't get into issues with estimating via multiplicative error only
#> ADD : 0.1 : Additive residual error
#>
#> [TABLE]
#> double IPRED = CENT/Vi;
#> double DV = CENT/Vi*(1+PROP) + ADD;
#>
#> [CAPTURE] @annotated
#> DV : plasma concentration (mg/L)
#> IPRED : Individual predicted plasma concentration (mg/L)
#> CLi : Individual Clearance (L/hr)
#> Vi : Individual Volume (L)one_cmt_iv %>%
model_details %>%
filter(block != "CAPTURE") %>%
kable()| block | name | descr | unit | options | value |
|---|---|---|---|---|---|
| PARAM | CL | Clearance | L/hr | . | 3.00 |
| PARAM | V | Volume | L | . | 35.00 |
| CMT | CENT | Central compartment | mg | . | 0.00 |
| OMEGA | nCL | Random effect on CL | . | . | 0.10 |
| OMEGA | nV | Random effect on V | . | . | 0.04 |
| SIGMA | PROP | Proportional error | . | . | 0.04 |
| SIGMA | ADD | Additive residual error | . | . | 0.10 |
simulated_data <- one_cmt_iv %>%
data_set(for_dosing) %>%
mrgsim(end = 24, delta = 0.25) %>% as_data_frame1.3 Distribution of peak and trough values
- ‘peak’ defined as 1 hr post infusion and trough 1 hour prior to when next dose would begin
simulated_data %>%
filter(TIME %in% c(2, 23)) %>%
mutate(DV = ifelse(DV < 0, 0, DV)) %>%
select(ID, DV) %>%
group_by(ID) %>%
summarize_all(funs(min, max)) %>%
gather(sample, value, -ID) %>%
ggplot(aes(x = value)) +
geom_density() +
facet_wrap(~sample, scales = "free") +
theme_bw() +
base_theme()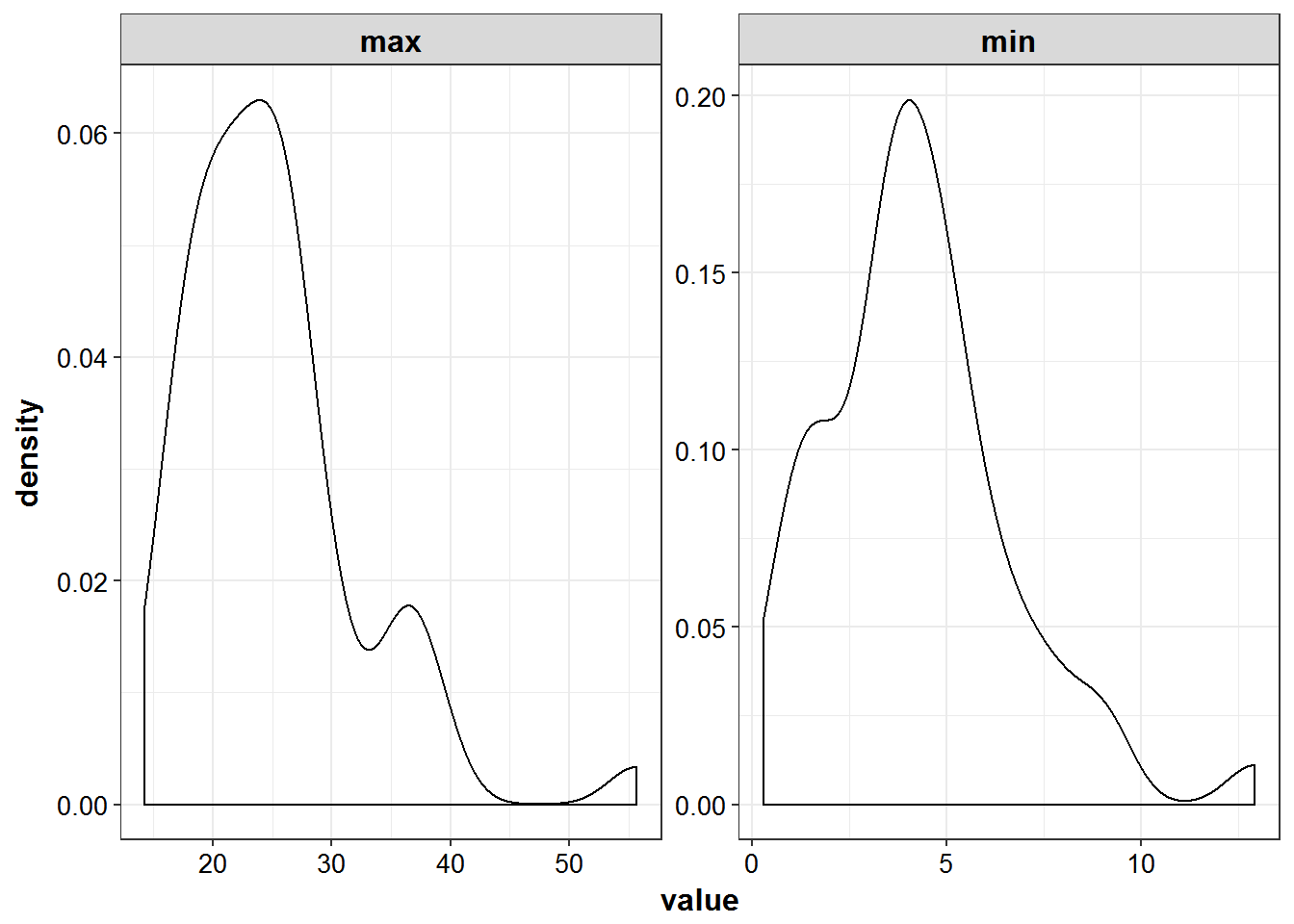
1.4 Predicted Profiles
simulated_data %>%
filter(IPRED > 0.1) %>%
ggplot(aes(x = TIME, y = IPRED, group = ID)) +
geom_line(size = 1.05, alpha = 0.8) + theme_bw() +
base_theme() + scale_y_log10()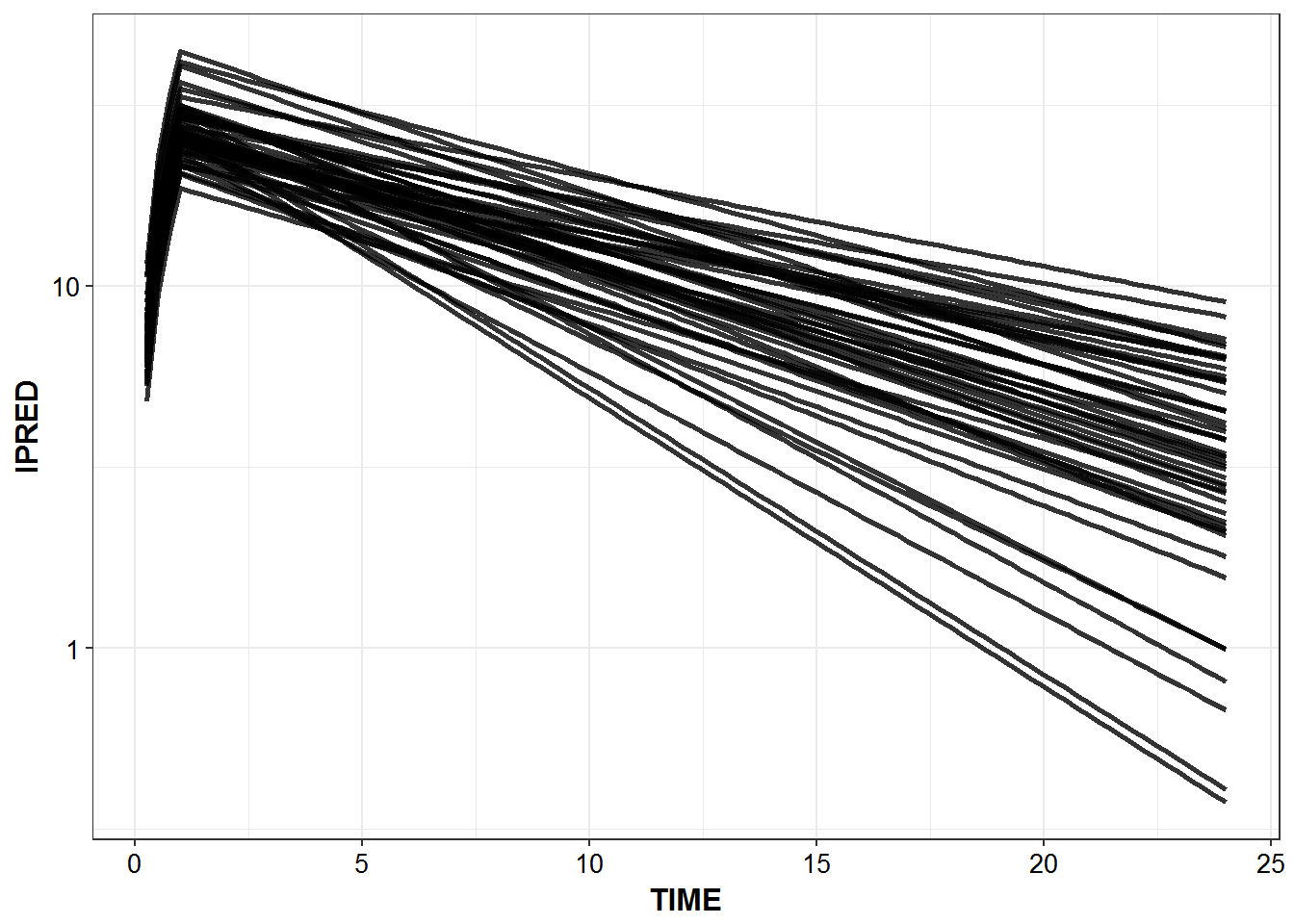
1.5 Real world sampling and LLOQ
sample_times <- c(2, 4, 8, 16, 24)
LLOQ <- 0.1
sampled_data <- simulated_data %>%
filter(TIME %in% sample_times, DV > LLOQ) Show which, if any, timepoints have bql values and determine percent bql
sampled_data %>%
count(TIME) %>%
mutate(baseline = first(n),
perc_bql = 100 - n/baseline*100) %>%
filter(perc_bql > 0) %>%
select(TIME, perc_bql) %>% knitr::kable()TIME perc_bql —– ———
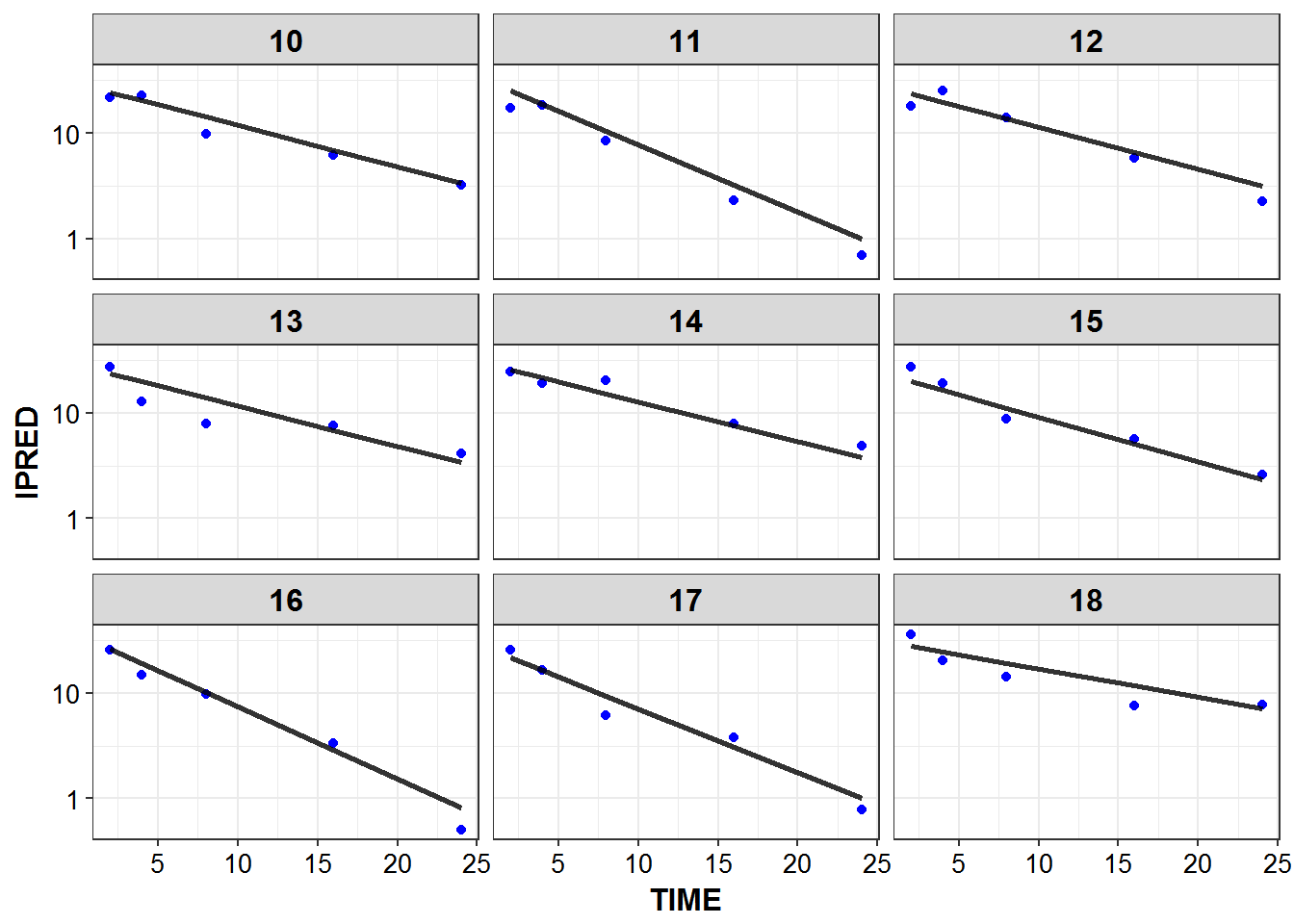
1.6 IPRED and DV vs TIME for all individuals at sampled times
list_plots <- sampled_data %>%
mutate(PNUM = ids_per_plot(ID)) %>%
split(.$PNUM) %>%
map(~
ggplot(., aes(x = TIME, y = IPRED, group = ID)) +
geom_point(aes(y = DV), color = "blue") +
geom_line(size = 1.05, alpha = 0.8) + theme_bw() +
base_theme() + facet_wrap(~ID) +
scale_y_log10()
)
print_plots(list_plots)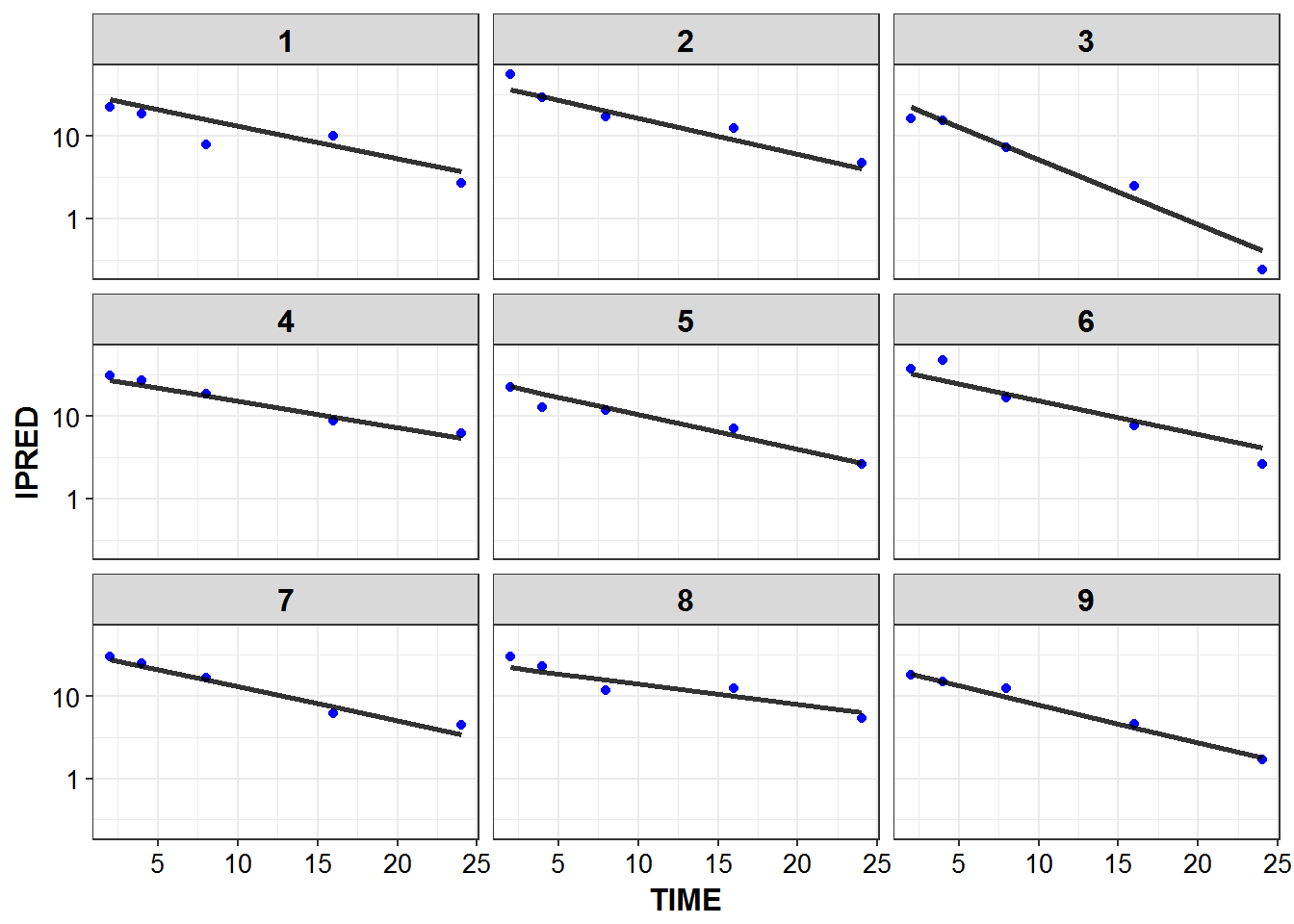
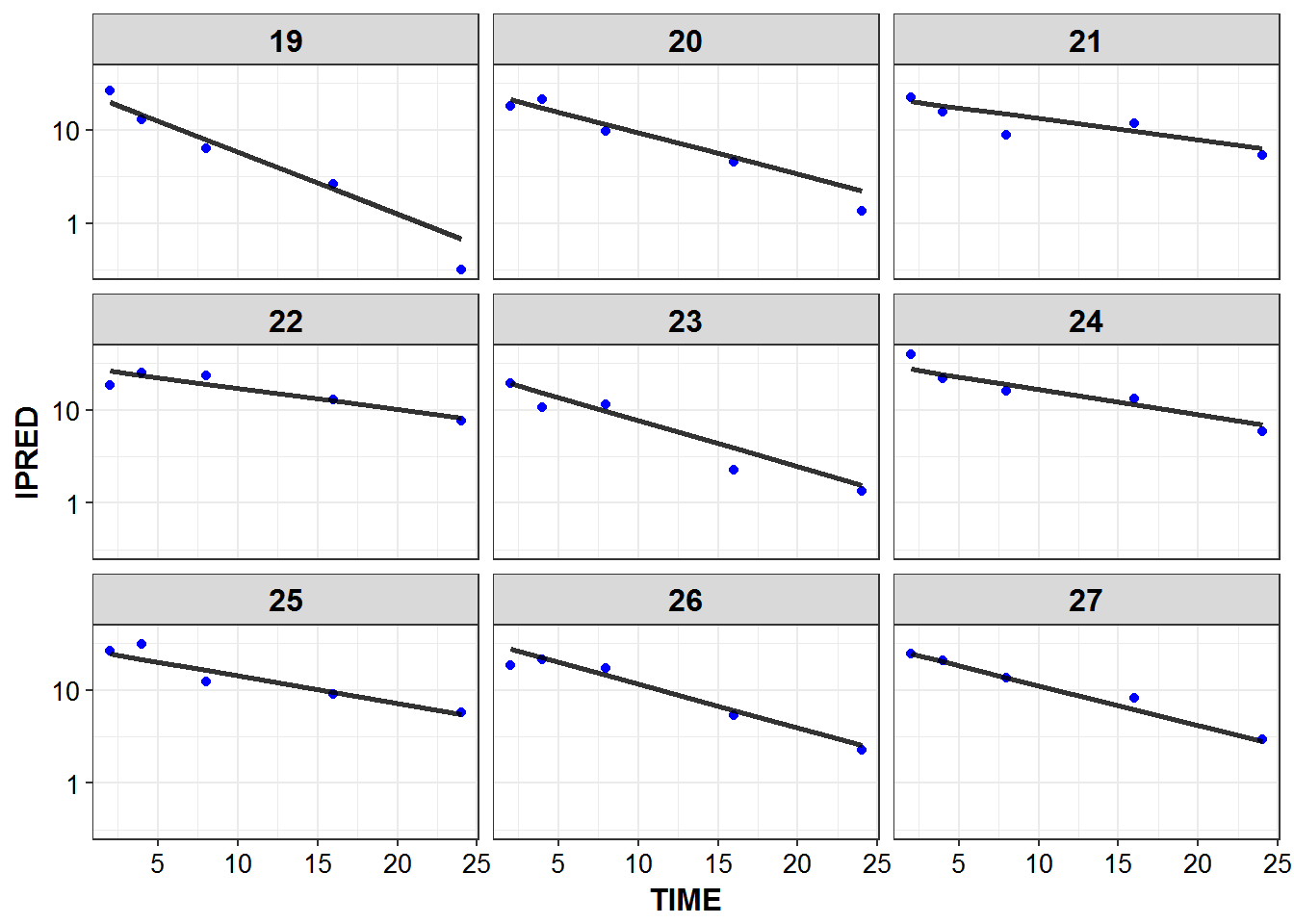
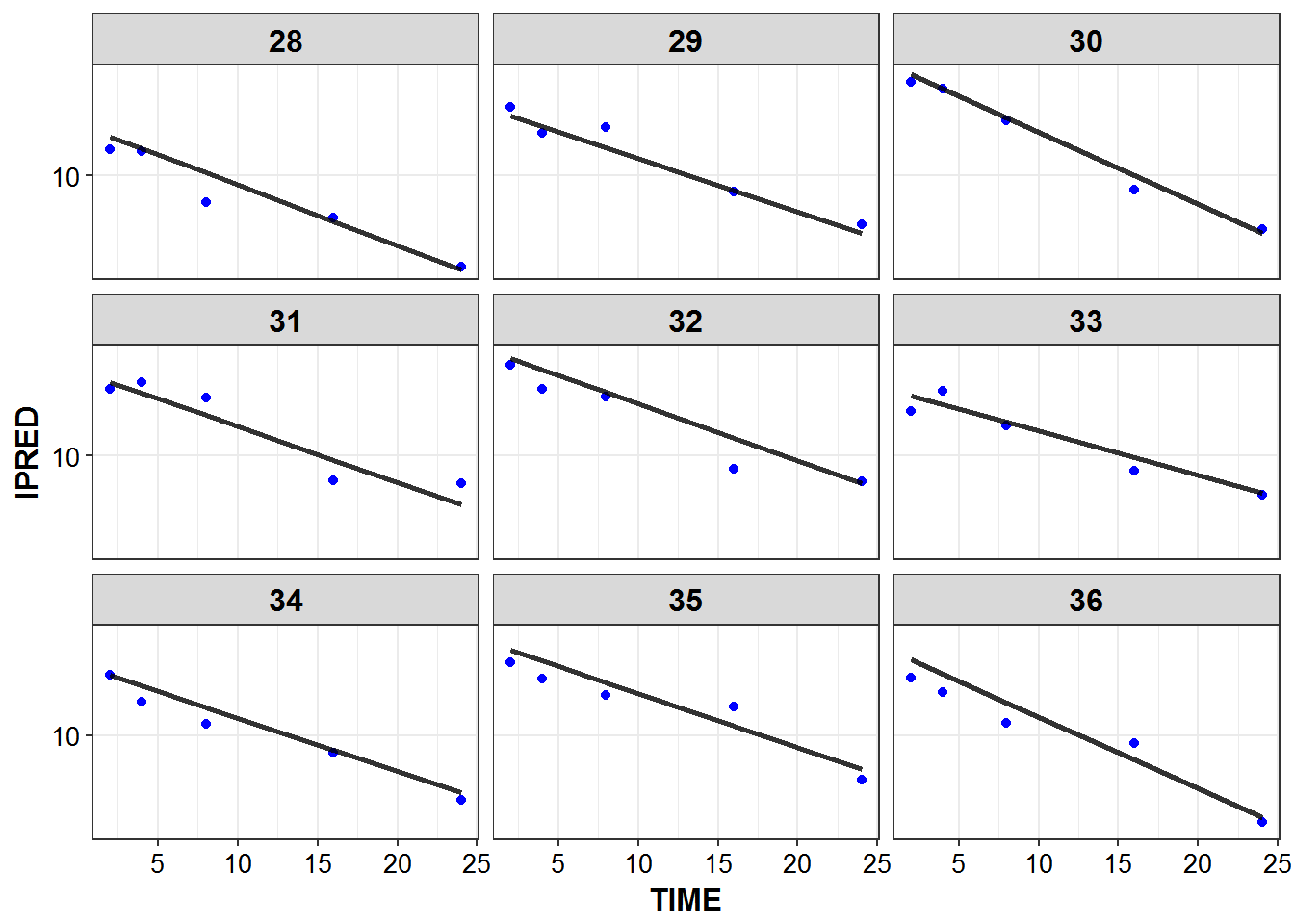
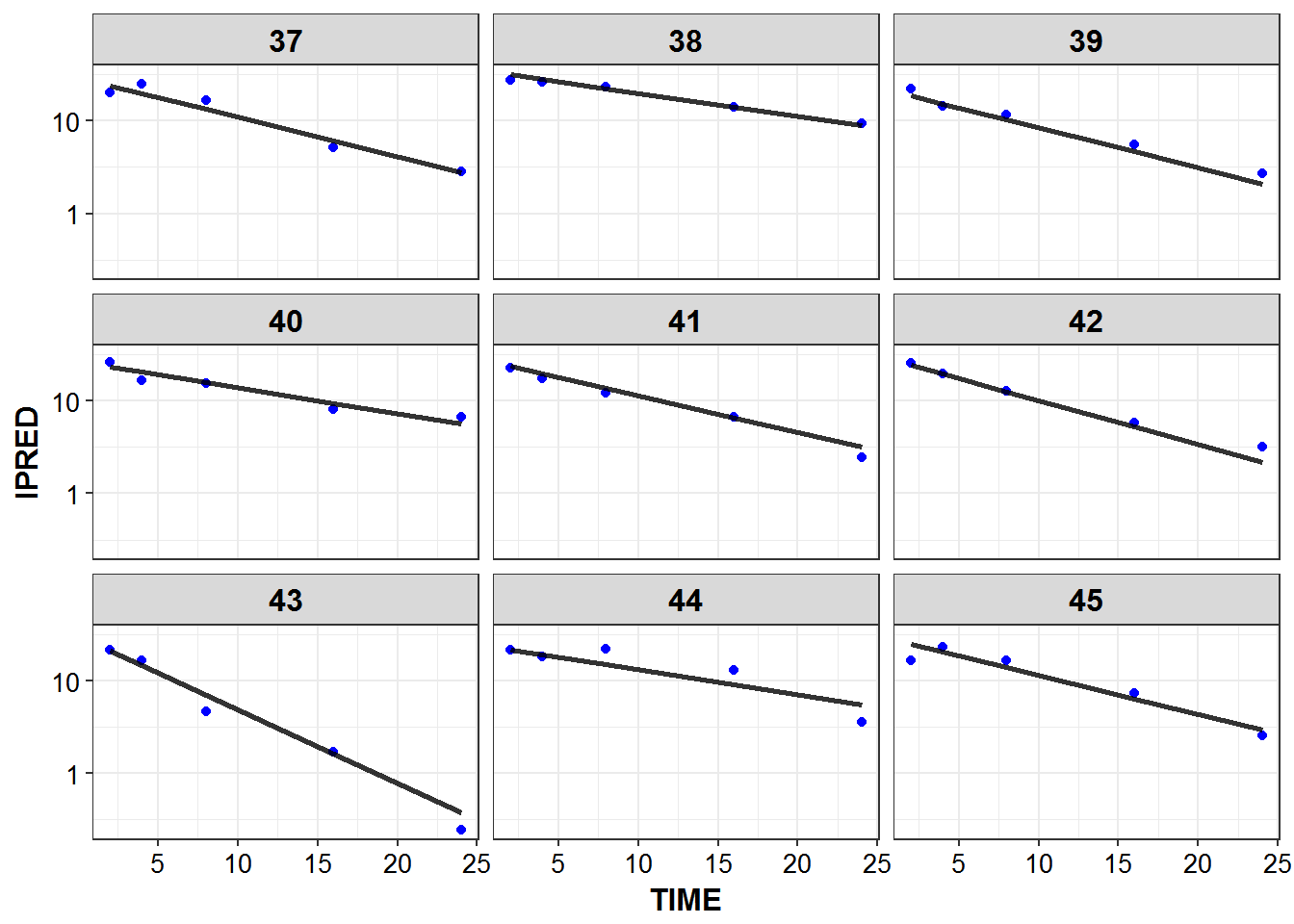
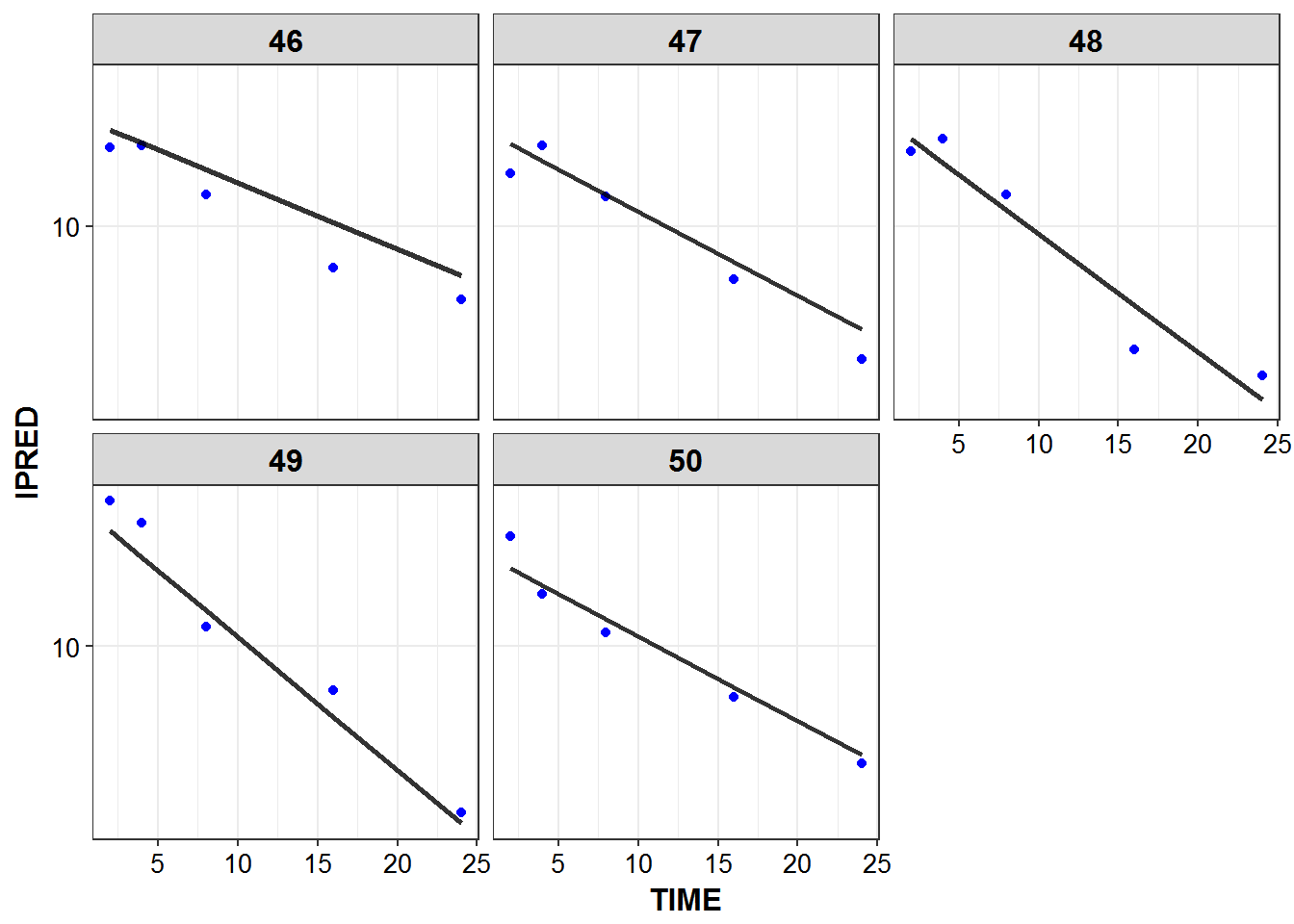
#> [[1]]
#> NULL
#>
#> [[2]]
#> NULL
#>
#> [[3]]
#> NULL
#>
#> [[4]]
#> NULL
#>
#> [[5]]
#> NULL
#>
#> [[6]]
#> NULL1.7 Prepare for nonmem
nm_dat <- sampled_data %>% select(ID, TIME, DV) %>%
mutate(CMT = 1,
EVID = 0
) %>%
bind_rows(for_dosing) %>%
arrange(ID, TIME, desc(EVID))kable(head(nm_dat))| ID | TIME | DV | CMT | EVID | AMT | RATE |
|---|---|---|---|---|---|---|
| 1 | 0 | NA | 1 | 1 | 1000 | 1000 |
| 1 | 2 | 22.40 | 1 | 0 | NA | NA |
| 1 | 4 | 18.59 | 1 | 0 | NA | NA |
| 1 | 8 | 7.88 | 1 | 0 | NA | NA |
| 1 | 16 | 10.20 | 1 | 0 | NA | NA |
| 1 | 24 | 2.70 | 1 | 0 | NA | NA |
write_nonmem(nm_dat, "../modeling/mdata/simple_nocovar_50id_6tp.csv")session_details <- devtools::session_info()
session_details$platform
#> setting value
#> version R version 3.3.2 (2016-10-31)
#> system x86_64, mingw32
#> ui RTerm
#> language (EN)
#> collate English_United States.1252
#> tz America/New_York
#> date 2016-12-13
knitr::kable(session_details$packages)| package | * | version | date | source |
|---|---|---|---|---|
| backports | 1.0.4 | 2016-10-24 | CRAN (R 3.3.2) | |
| bookdown | 0.2 | 2016-11-12 | CRAN (R 3.3.2) | |
| devtools | 1.12.0 | 2016-06-24 | CRAN (R 3.3.2) | |
| digest | 0.6.10 | 2016-08-02 | CRAN (R 3.3.2) | |
| evaluate | 0.10 | 2016-10-11 | CRAN (R 3.3.2) | |
| htmltools | 0.3.5 | 2016-03-21 | CRAN (R 3.3.2) | |
| httpuv | 1.3.3 | 2015-08-04 | CRAN (R 3.3.2) | |
| knitr | 1.15 | 2016-11-09 | CRAN (R 3.3.2) | |
| magrittr | 1.5 | 2014-11-22 | CRAN (R 3.3.2) | |
| memoise | 1.0.0 | 2016-01-29 | CRAN (R 3.3.2) | |
| mime | 0.5 | 2016-07-07 | CRAN (R 3.3.2) | |
| miniUI | 0.1.1 | 2016-01-15 | CRAN (R 3.3.2) | |
| R6 | 2.2.0 | 2016-10-05 | CRAN (R 3.3.2) | |
| Rcpp | 0.12.8 | 2016-11-17 | CRAN (R 3.3.2) | |
| rmarkdown | 1.2 | 2016-11-21 | CRAN (R 3.3.2) | |
| rprojroot | 1.1 | 2016-10-29 | CRAN (R 3.3.2) | |
| shiny | 0.14.2 | 2016-11-01 | CRAN (R 3.3.2) | |
| stringi | 1.1.2 | 2016-10-01 | CRAN (R 3.3.2) | |
| stringr | 1.1.0 | 2016-08-19 | CRAN (R 3.3.2) | |
| withr | 1.0.2 | 2016-06-20 | CRAN (R 3.3.2) | |
| xtable | 1.8-2 | 2016-02-05 | CRAN (R 3.3.2) | |
| yaml | 2.1.13 | 2014-06-12 | CRAN (R 3.3.2) |