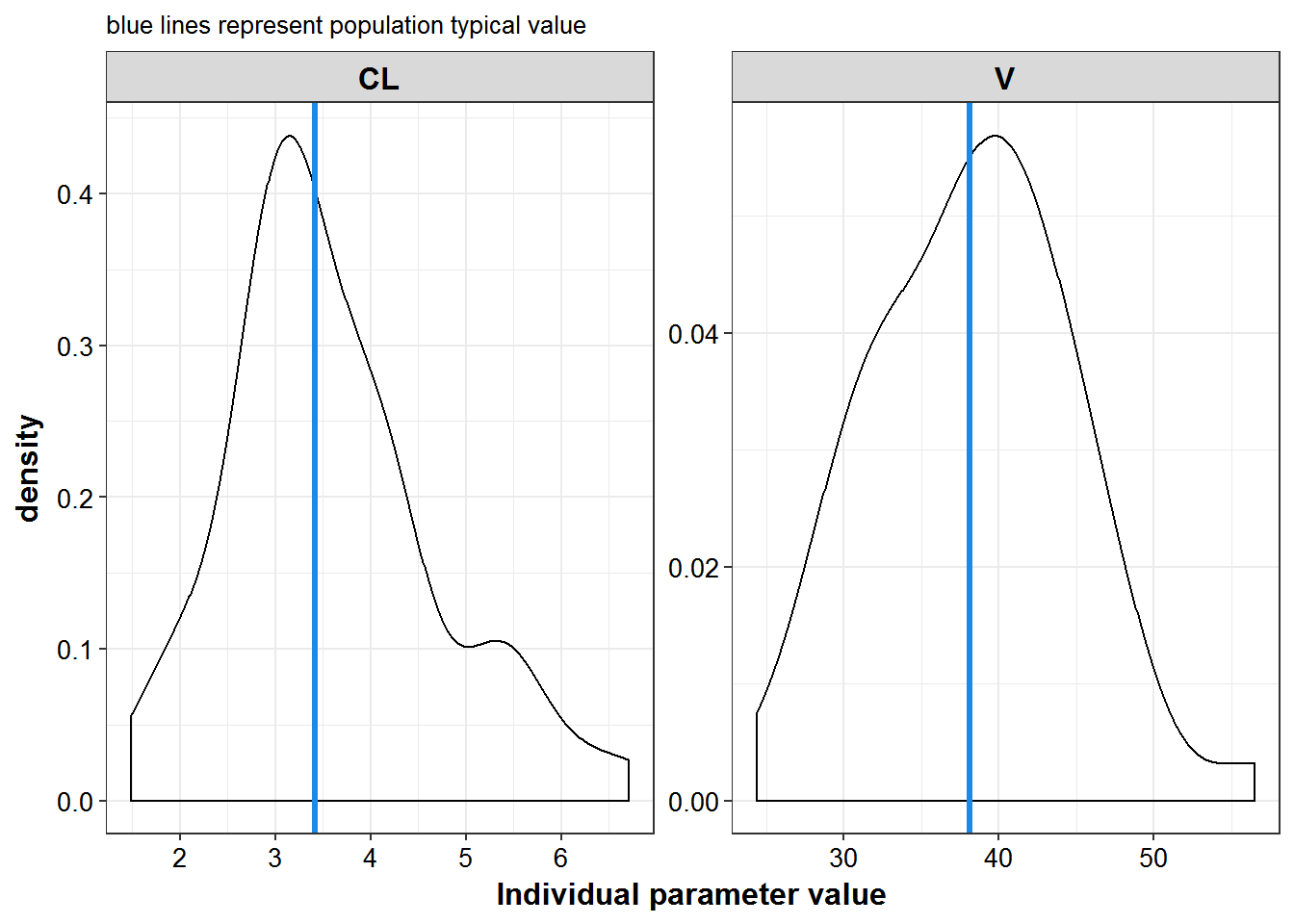
Parameter values
patab1 %>%
select(ID:TVV) %>%
gather(iparam, value, CL, V) %>%
mutate(TVP = ifelse(iparam == "CL", TVCL, TVV)) %>%
ggplot(aes(x = value, group = iparam)) +
geom_density() +
geom_vline(aes(xintercept = TVP), color = mcp$blue, size = 1.2) +
facet_wrap(~iparam, scales = "free") + theme_bw() +
base_theme() +
labs(x = "Individual parameter value", subtitle="blue lines represent population typical value")
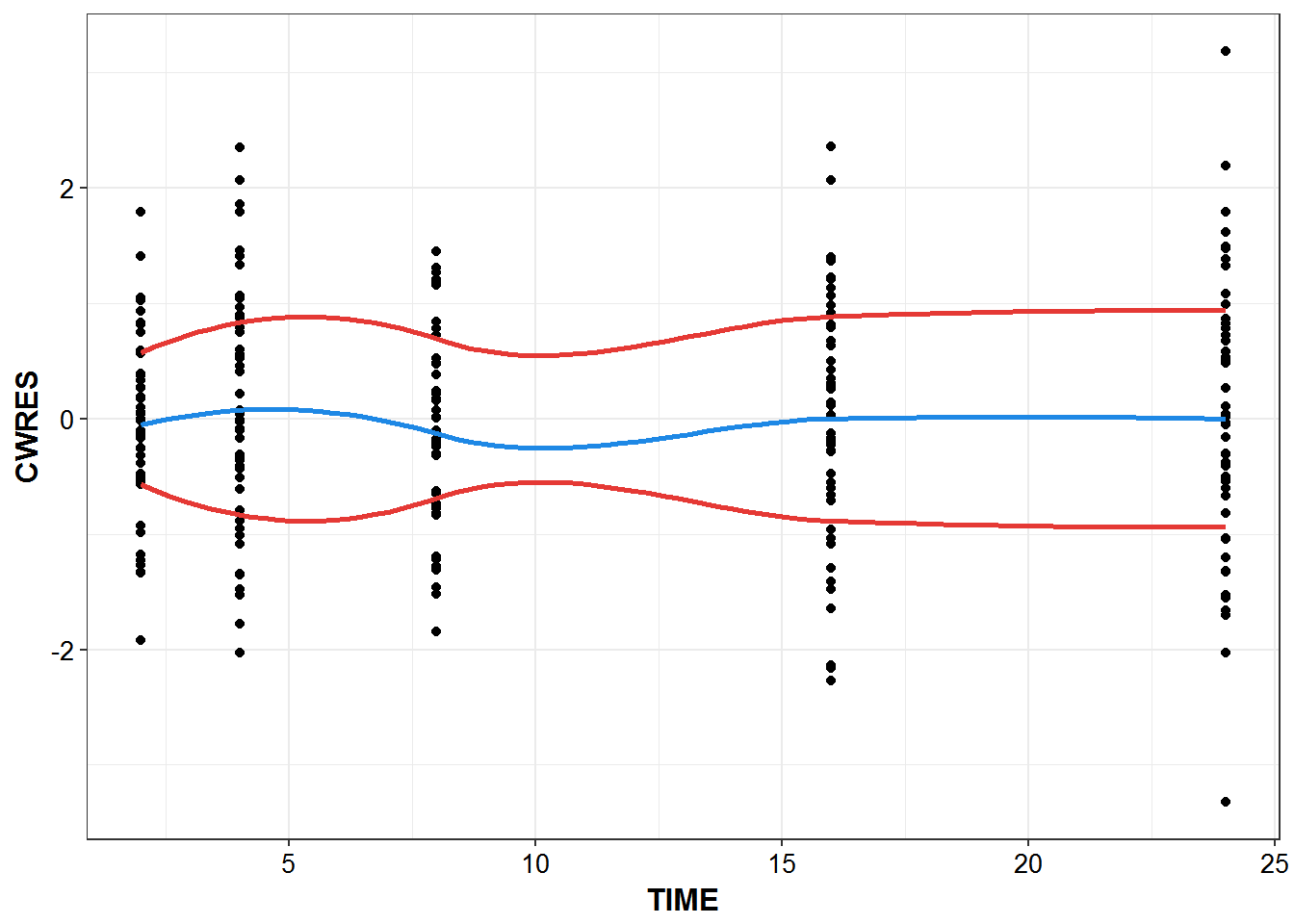
Individual diagnostic plots
sdtab1 %>%
filter(TIME >0) %>%
ggplot(aes(x = TIME, y = CWRES)) + geom_point() +
geom_smooth(se = FALSE, color = mcp$blue) +
theme_bw() +
base_theme() +
geom_smooth(aes(y = abs(CWRES)), se = F, color = mcp$red) +
geom_smooth(aes(y = -abs(CWRES)), se = F, color = mcp$red)
#> `geom_smooth()` using method = 'loess'
#> `geom_smooth()` using method = 'loess'
#> `geom_smooth()` using method = 'loess'
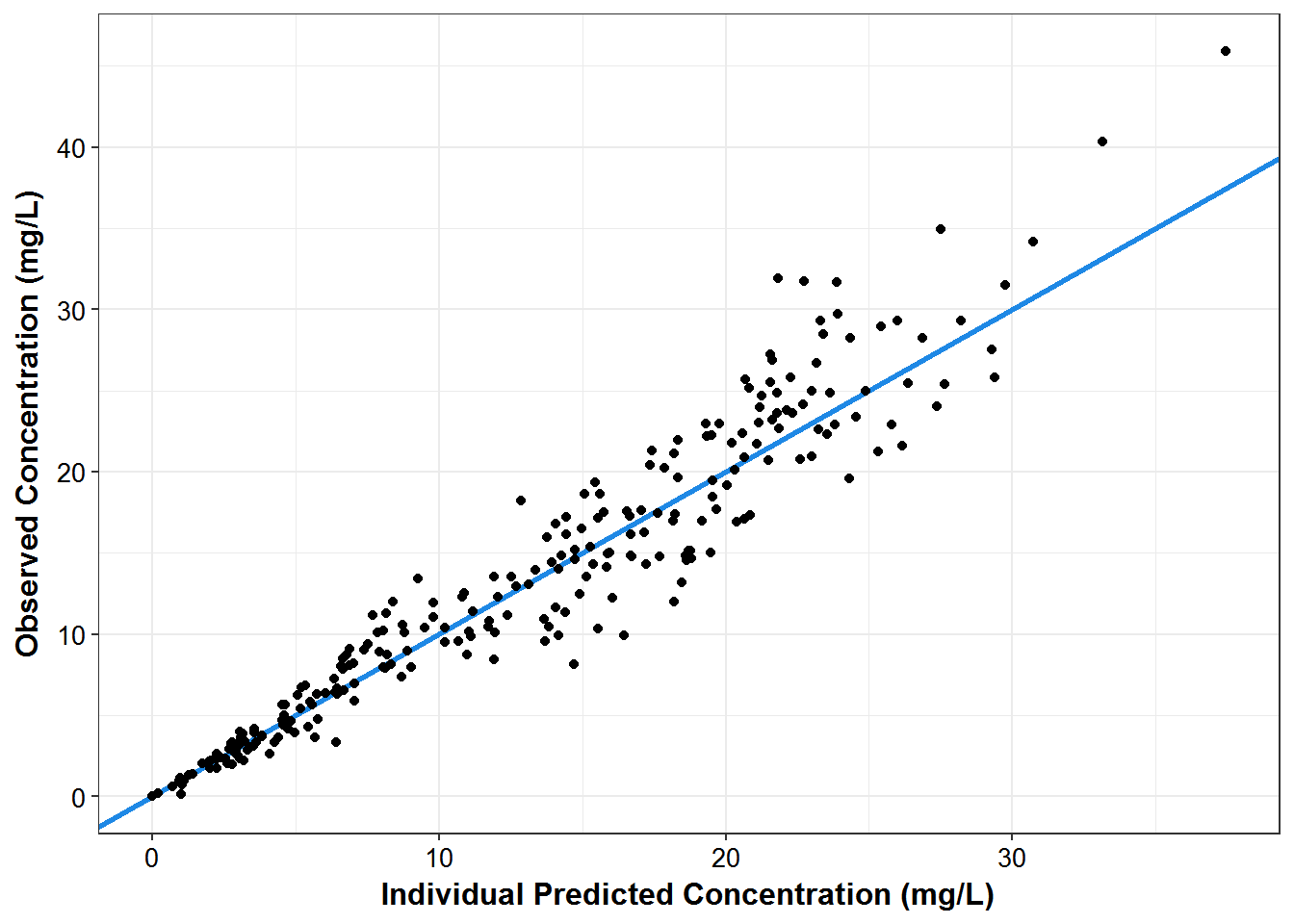
sdtab1 %>%
ggplot(aes(x = IPRED, y = DV)) +
geom_abline(color = mcp$blue, size = 1.1) +
geom_point() +
theme_bw() +
base_theme() +
labs(x = "Individual Predicted Concentration (mg/L)",
y = "Observed Concentration (mg/L)")
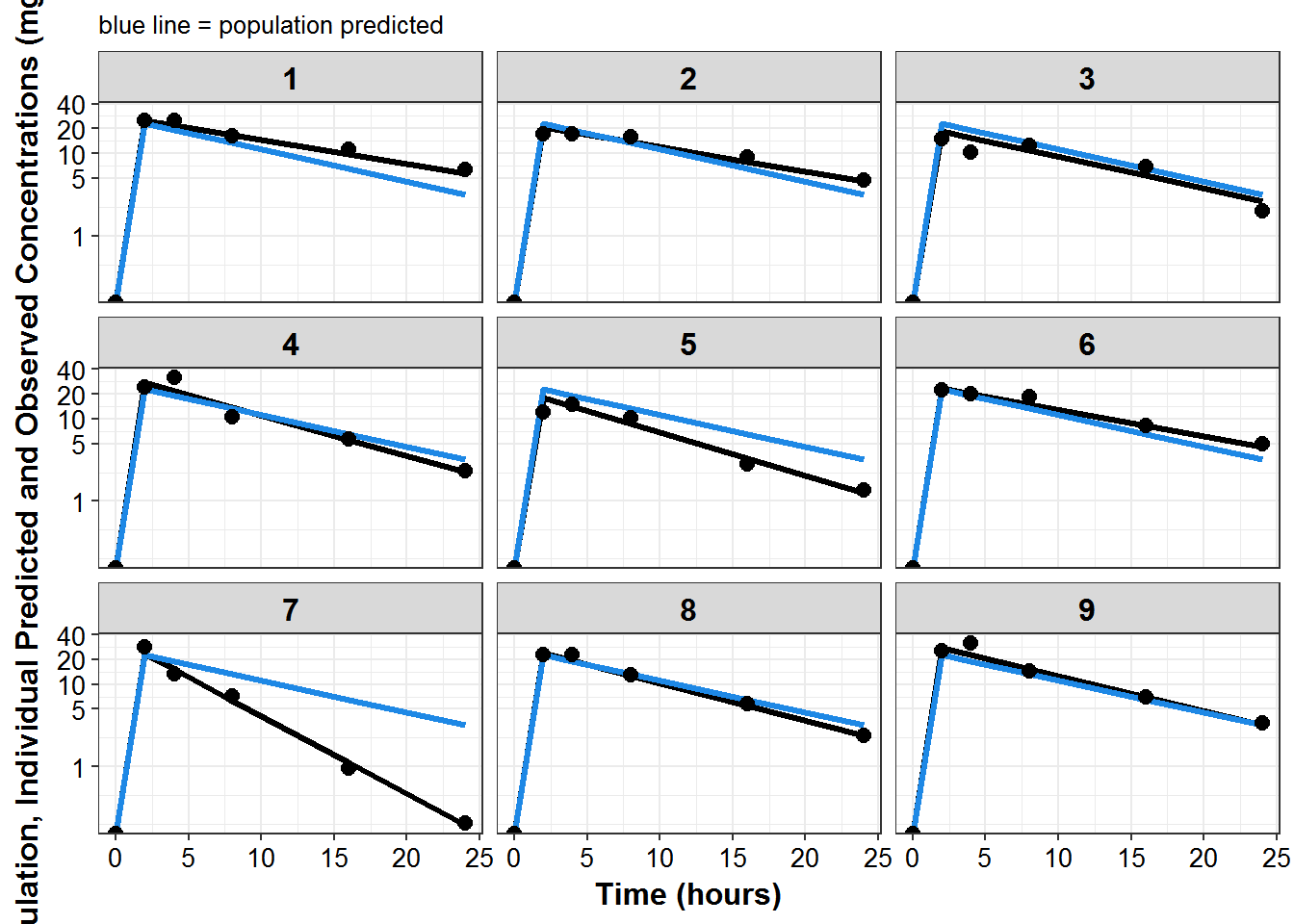
plot_list <- sdtab1 %>%
mutate(PNUM = ids_per_plot(ID)) %>%
split(.$PNUM) %>%
map(~
ggplot(., aes(x = TIME, y = IPRED, group = ID)) +
geom_line(size = 1.1) +
geom_line(aes(y = PRED), size = 1.1, color = mcp$blue) +
geom_point(aes(y = DV), size = 2.5) + facet_wrap(~ID) +
theme_bw() +
base_theme() +
labs(y = "Population, Individual Predicted and Observed Concentrations (mg/L)",
x = "Time (hours)",
subtitle = "blue line = population predicted"
) +
scale_y_log10(breaks = c(1, 5, 10, 20, ceiling(max(.$DV)/10)*10))
)
print_plots(plot_list)
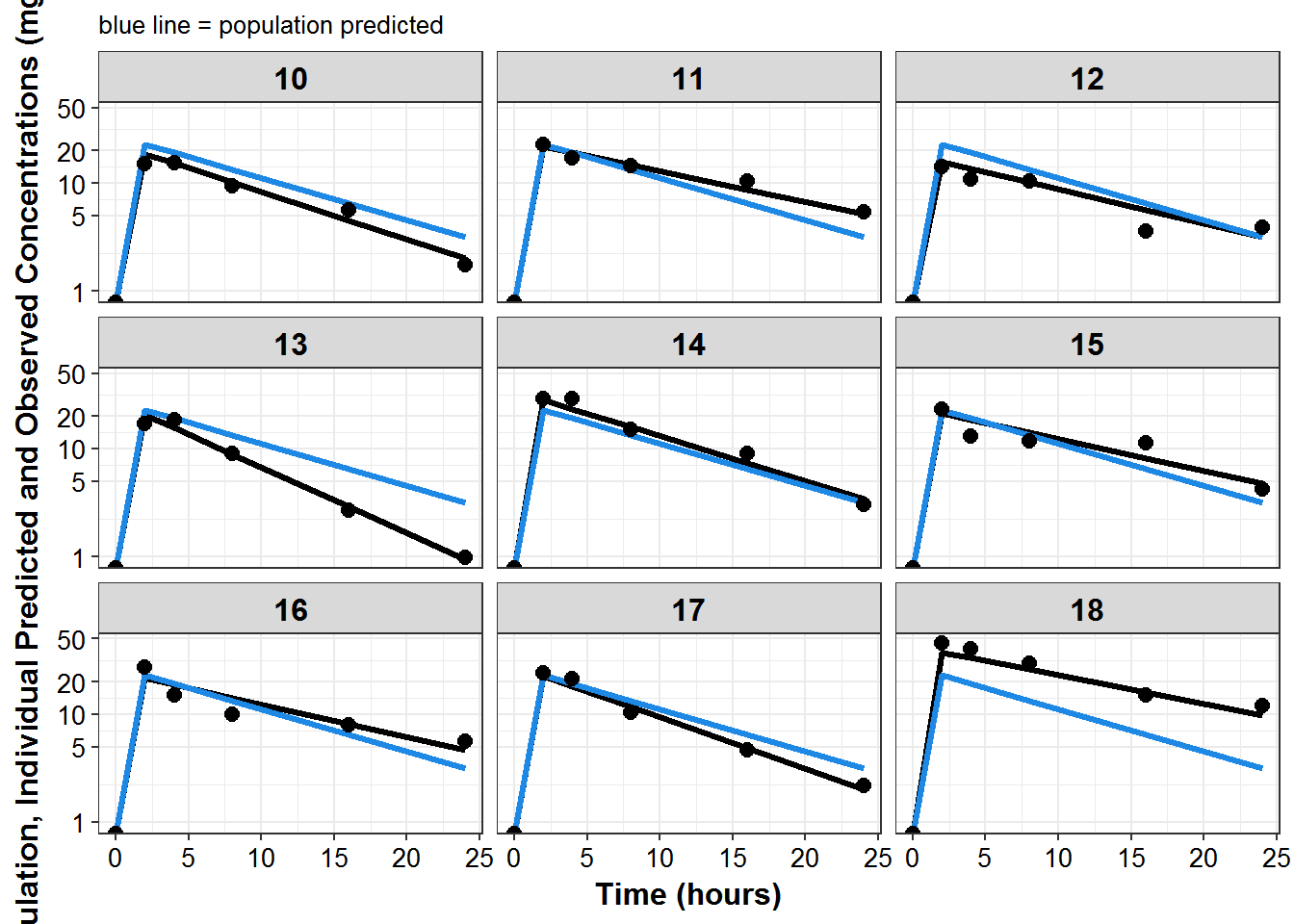
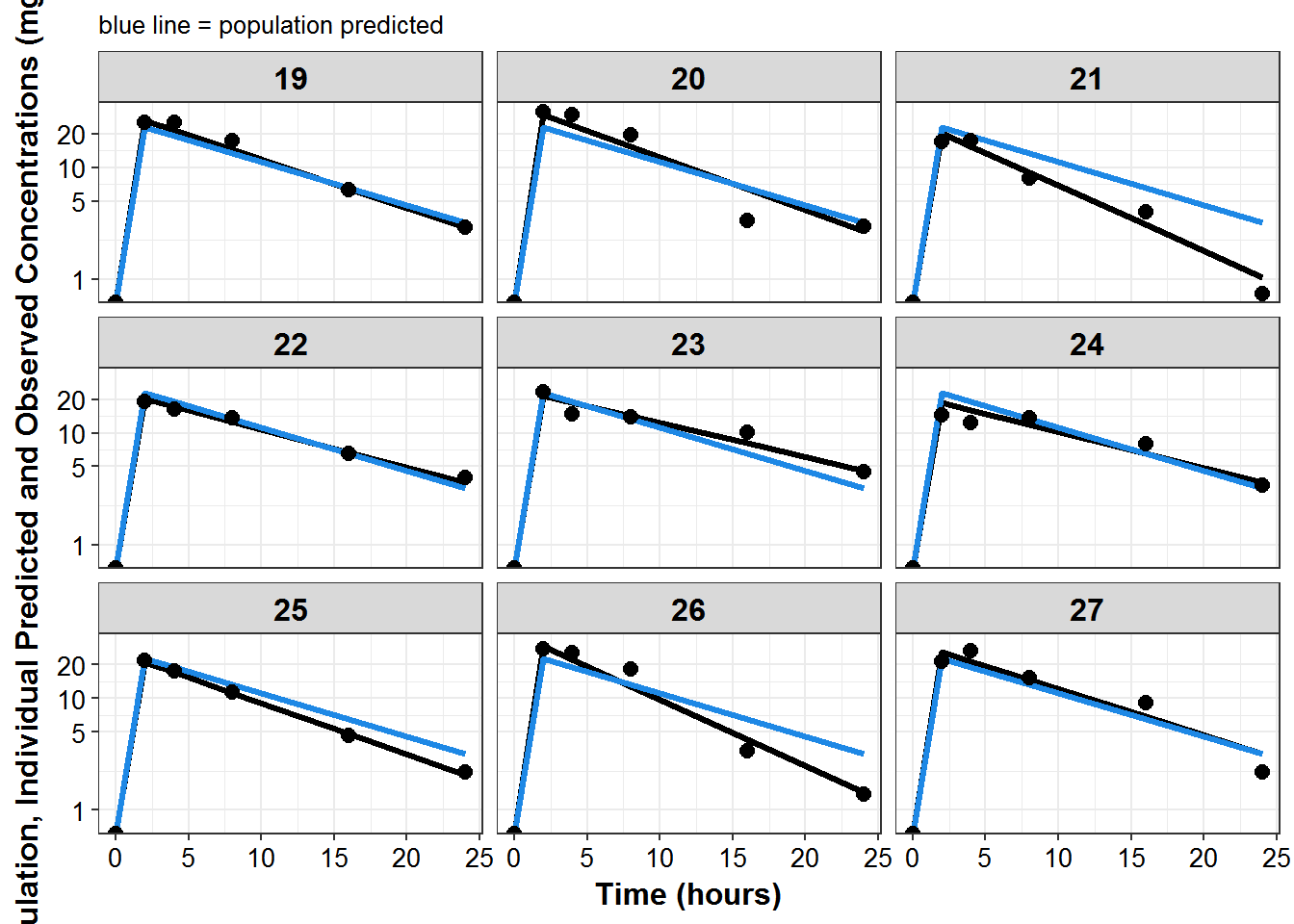
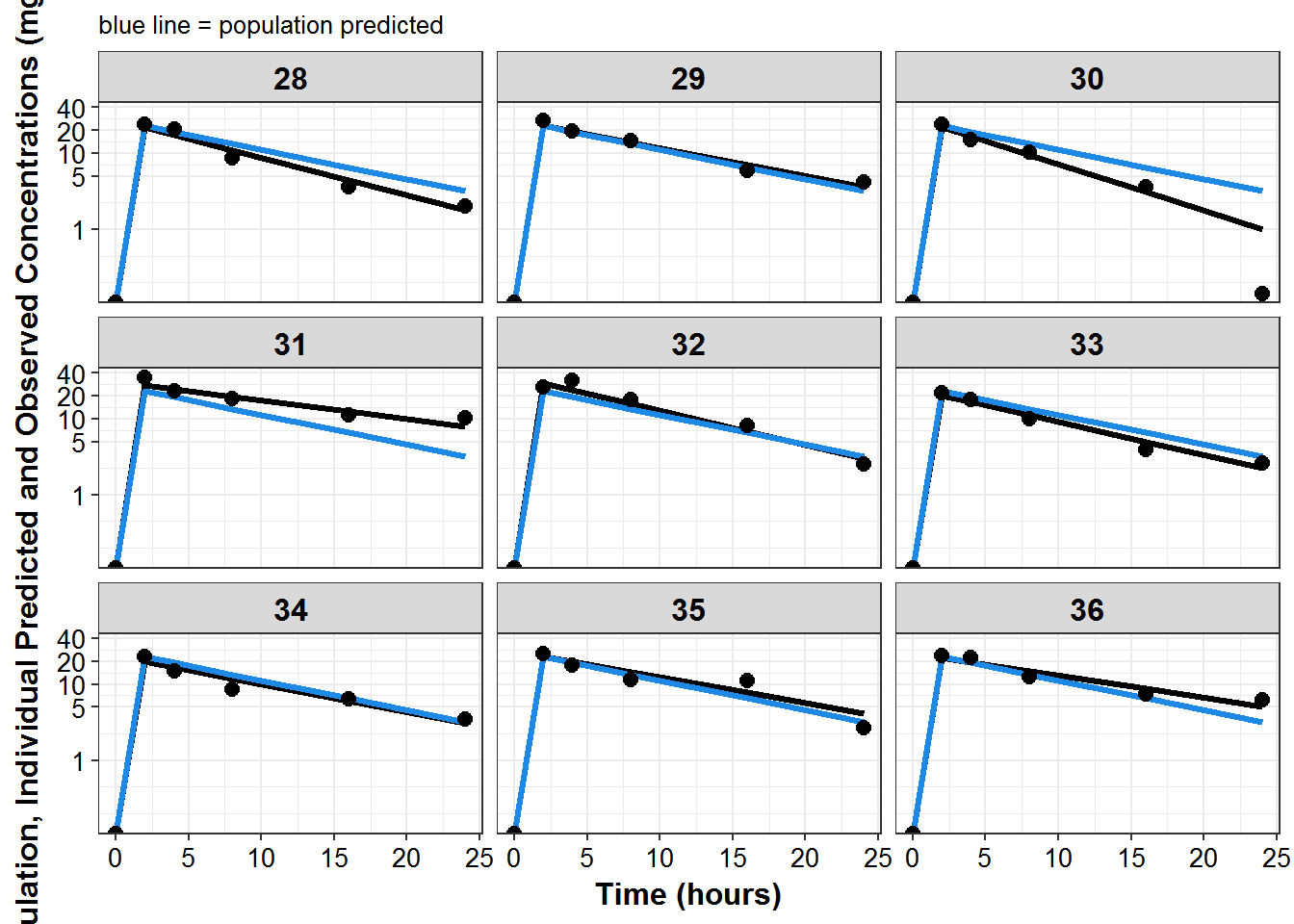
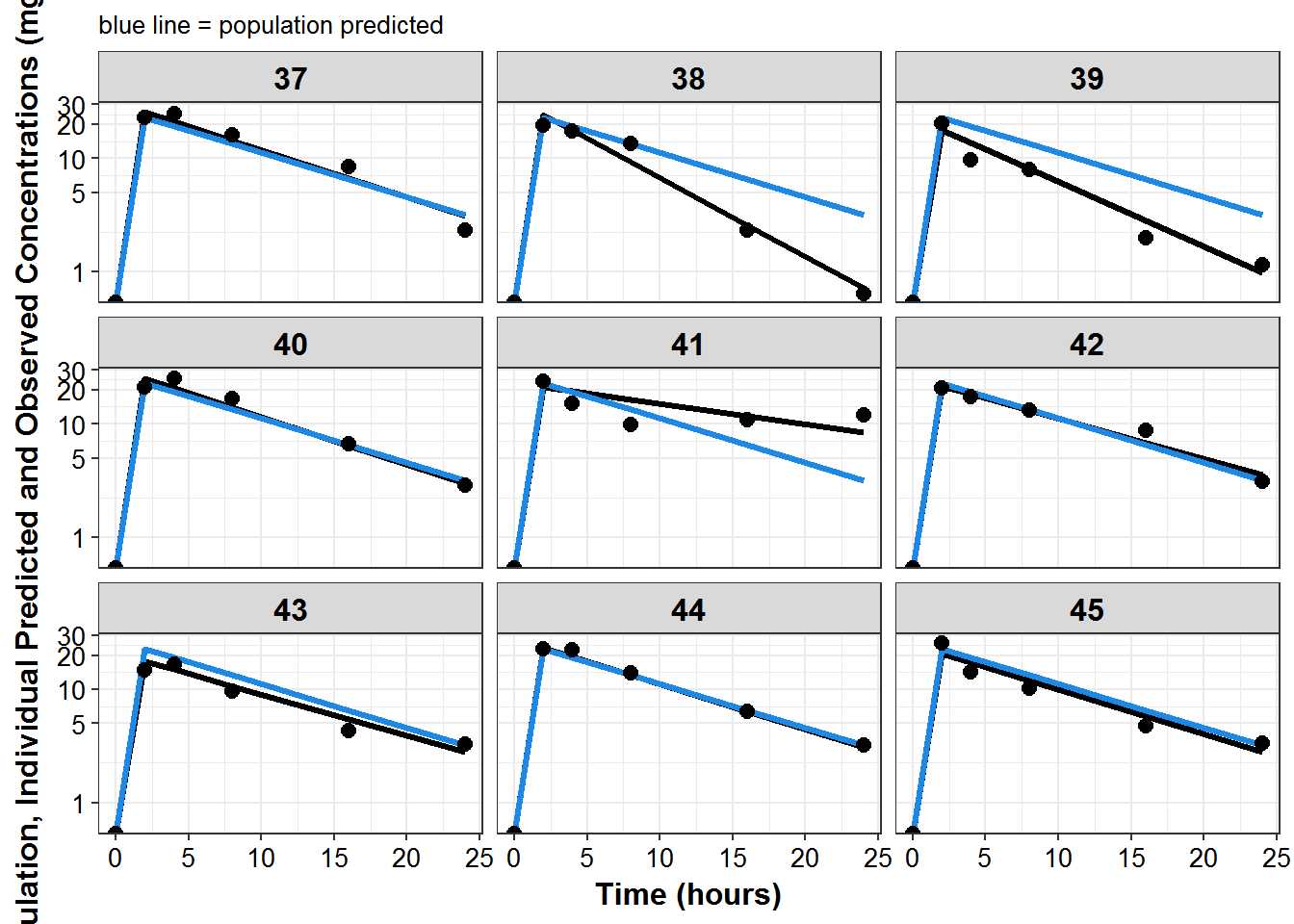

#> [[1]]
#> NULL
#>
#> [[2]]
#> NULL
#>
#> [[3]]
#> NULL
#>
#> [[4]]
#> NULL
#>
#> [[5]]
#> NULL
#>
#> [[6]]
#> NULL
session_details <- devtools::session_info()
session_details$platform
#> setting value
#> version R version 3.3.2 (2016-10-31)
#> system x86_64, mingw32
#> ui RTerm
#> language (EN)
#> collate English_United States.1252
#> tz America/New_York
#> date 2016-12-13
knitr::kable(session_details$packages)
| backports |
|
1.0.4 |
2016-10-24 |
CRAN (R 3.3.2) |
| bookdown |
|
0.2 |
2016-11-12 |
CRAN (R 3.3.2) |
| devtools |
|
1.12.0 |
2016-06-24 |
CRAN (R 3.3.2) |
| digest |
|
0.6.10 |
2016-08-02 |
CRAN (R 3.3.2) |
| evaluate |
|
0.10 |
2016-10-11 |
CRAN (R 3.3.2) |
| htmltools |
|
0.3.5 |
2016-03-21 |
CRAN (R 3.3.2) |
| httpuv |
|
1.3.3 |
2015-08-04 |
CRAN (R 3.3.2) |
| knitr |
|
1.15 |
2016-11-09 |
CRAN (R 3.3.2) |
| magrittr |
|
1.5 |
2014-11-22 |
CRAN (R 3.3.2) |
| memoise |
|
1.0.0 |
2016-01-29 |
CRAN (R 3.3.2) |
| mime |
|
0.5 |
2016-07-07 |
CRAN (R 3.3.2) |
| miniUI |
|
0.1.1 |
2016-01-15 |
CRAN (R 3.3.2) |
| R6 |
|
2.2.0 |
2016-10-05 |
CRAN (R 3.3.2) |
| Rcpp |
|
0.12.8 |
2016-11-17 |
CRAN (R 3.3.2) |
| rmarkdown |
|
1.2 |
2016-11-21 |
CRAN (R 3.3.2) |
| rprojroot |
|
1.1 |
2016-10-29 |
CRAN (R 3.3.2) |
| shiny |
|
0.14.2 |
2016-11-01 |
CRAN (R 3.3.2) |
| stringi |
|
1.1.2 |
2016-10-01 |
CRAN (R 3.3.2) |
| stringr |
|
1.1.0 |
2016-08-19 |
CRAN (R 3.3.2) |
| withr |
|
1.0.2 |
2016-06-20 |
CRAN (R 3.3.2) |
| xtable |
|
1.8-2 |
2016-02-05 |
CRAN (R 3.3.2) |
| yaml |
|
2.1.13 |
2014-06-12 |
CRAN (R 3.3.2) |








